ICACSIS 2014
2014 lnternotionol Conference on
Advanced Con7puter Science one!
infor1notion
sセカウエ・ョQウ@
October 18th and 19th 2014
Ambhara Hotel, Blok M
Jakarta, Indonesia
ISBN : 978-979-1421-22-5
+.IEEE
INOONESIA
SECTION
ICACSIS
2014
2014 International Conference on
Advanced Computer Science and Information Systems (Proceedings)
plenary and invited speakers.
Welcome Message from
General Chairs
This international conference is organized by the Faculty of Computer Science, Universitas Indonesia, and is intended to be the first step towards a top class conference on Computer Science and Information Systems. We believe that this international conference will give opportunities for sharing and exchanging original research ideas and opinions, gaining inspiration for future research, and broadening knowledge about various fields in advanced compi.ner science and information systems, amongst members of Indonesian research communities, together with researchers from Germany, Singapore, Thailand, France, Algeria, Japan, Malaysia, Philippines, United Kingdom, Sweden, United States and other countries.
This conference focuses on the development of computer science and information systems. Along with 4 plenary and 2 invited speeches, the proceedings of this conference contains 71 papers which have been selected from a total of 132 papers from twelve different countries. These selected papers will be presented during the conference.
We also want to express our sincere appreciation to the members of the Program Committee for their critical review of the submitted papers, as well as the Organizing Committee for the time and energy they have devoted to editing the proceedings and arranging the logistics of holding this conference. We would also like to give appreciation to the authors who have submitted their excellent works to this conference. Last but not least, we would like to extend our gratitude to the Ministry of Education of the Republic of Indonesia, the Rector of Universitas Indonesia, Universitas Tarumanagara, Bogor Agricultural Institute, and the Dean of the Faculty of Computer Science for their continued support towards the !CACS!S 2014 conference.
Sincerely yours,
General Chairs
Welcome Message from
The Dean of Faculty of Computer Science, Universitas
Indonesia
On behalf of all the academic staff and students of the Faculty of Computer Science, Universitas Indonesia, I would like to extend our warmest welcome to all the participants to the Ambhara Hotel, Jakarta on the occasion of the 2014 International Conference on Advanced Computer Science and Information Systems (ICACSIS).
Just like the previous five events in this series (ICACSIS 2009, 2010, 2011, 2012, and 2013), I am confident that !CASIS 2014 will play an important role in encouraging activities in research and development of computer science and information technology in Indonesia, and give an excellent opportunity to forge collaborations between research institutions both within the country and with international partners. The broad scope of this event, which includes both theoretical aspects of computer science and practical, applied experience of developing information systems, provides a unique meeting ground for researchers spanning the whole spectrum of our discipline. 1 hope that over the next two days, some fruitful collaborations can be established.
I also hope that the special attention devoted this year to the field of pervasive computing, including the very exciting area of wireless sensor networks, will ignite the development of applications in this area to address the various needs of Indonesia's development.
I would like to express my sincere gratitude to the distinguished invited speakers for their presence and contributions to the conference. 1 also thank all the program committee members for their efforts in ensuring a rigorous review process to select high quality papers.
Finally, I sincerely hope that all the participants will benefit from the technical contents of this conference, and wish you a very successful conference and an enjoyable stay in Jakarta.
Sincerely,
Mirna Adriani, Dra, Ph.D.
Dean of the Faculty of Computer Science Universitas Indonesia
. .
. j,fa.j,1/fi,JJ4•@¥\,V,i
ICACS/S 2014COMMITTEES
Honorary Chairs:
• A. Jain, IEEE Fellow, Michigan State University, US • T. Fukuda, IEEE Fellow, Nagoya University, JP • M. Adriani, Universitas Indonesia, ID
General Chairs:
• E. K. Budiarjo, Universitas Indonesia, ID • D. I. Sensuse, Universitas Indonesia, ID • Z. A. Hasibuan, Universitas Indonesia, ID
Program Chairs:
• H. B. Santoso, Universitas Indonesia, ID • W. Jatmiko, Universitas Indonesia, ID
• A. Buono, Institut Pertanian Bogar, ID
• D. E. Herwindiati, Universitas Tarumanegara, ID
Section Chairs:
• K. Wastuwibowo, IEEE Indonesia Section, ID
Publication Chairs:
• A. Wibisono, Universitas Indonesia, ID
Program Committees:
•
A. Azurat, Universitas Indonesia, ID•
•
A. Fanar, Lembaga Ilmu Pengetahuan Indonesia, ID A. Kistijantoro, Institut Teknologi Bandung, ID
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
•
A. Purwarianti, Institut Teknologi Bandung, ID A. Nugroho, PTIK BPPT, ID
A. Srivihok, Kasetsart University, TH
A. Arifin Institut Teknologi Sepuluh Nopember, ID A. M. Arymurthy, Universitas Indonesia, ID A. N. Hidayanto, Universitas Indonesia, ID B. Wijaya, Universitas Indonesia, ID
B. Yuwono, Universitas Indonesia, ID B. Hardian, Universitas Indonesia, ID
B. Purwandari, Universitas Indonesia, ID B. A. Nazief, Universitas Indonesia, ID B. H. Widjaja, Universitas Indonesia, ID Denny, Universitas Indonesia, ID D. Jana, Computer Society of India, IN E. Gaura, Coventry University, UK E. Seo, Sungkyunkwan University, KR F: Gaol, IEEE Indonesia Section, ID
• M. I. Suryani, Universitas Indonesia, ID • M. Soleh, Universitas Indonesia, ID • M. Pravitasari, Universitas Indonesia, ID • N. Rahmah, Universitas Indonesia, ID • Putut, Universitas Indonesia, ID • R. Musfikar, Universitas Indonesia, ID • R. Latifah, Universitas Indonesia, ID • R. P. Rangkuti, Universitas Indonesia, ID • V. Ayumi, Universitas Indonesia, ID
...
1,12.fJ&i·d}M;{"fl
1cAcs1s
2014CONFERENCE INFORMATION
Dates
Organizer
Venue
Official language
Secretariat
October 18'" (Saturday) - October 19'" (Sunday) 2014
Faculty of Computer Science, Universitas Indonesia
Department of Computer Science, lnstitut Pertanian Bogor
Faculty of Information Technology, Universitas Tarumanegara
Ambhara Hotel
Jalan lskandarsyah Raya No. 1, Jakarta Selatan, OKI Jakarta, 12160, Indonesia
Phone : +62-21-2700 888
Fax : +62-21-2700 215
Website : http://www.ambharahotel.com/
English
Faculty of Computer Science, Universitas Indonesia
Kampus UI Depok
Depok, 16424
Indonesia
T: +62 21786 3419 ext. 3225
F: +62 21 786 3415
W: http://www.cs.ui.ac.id
Conference Website http://icacsis.cs.ui.ac.id
Sunday, October 19th, 2014-CONFERENCE
Time Event Event Details Rooms
08.00-09.00 Registration
Ors. Harry Waluyo, M.Hum
from Directorate General of Media,
09.00-10.00 Plenary Speech Ill Design, Science & Technology Based
Creative Economy Dirgantara Room,
2"' Floor
10.00-10.lS Coffee Break
Prof. Masatoshi Ishikawa
10.15-11.30 Plenary Speech IV from University of Tokyo, JP
11.30-12.30 Lunch
Parallel Session IV: See Technical (Parallel Session IV Elang, Kasuari,
12.30-14.00
Four Parallel Sessions Schedule) Merak, Cendrawasih
Room, Lobby Level
Parallel Session V: See Technical (Parallel Session V Elang, Kasuari,
14.00-15.30
Four Parallel Sessions Schedule) Merak, Cendrawasih
Room, Lobby Level
15.30-16.00 Coffee Break
16.00-16.30 Closing Ceremony Awards Announcement and Photo Dirgantara Room,
Session 2"' Floor
Table of Contents
Welcome Message from General Chairs
Welcome Message from Dean of Faculty of Computer Science University of Indonesia
Committees
Conference Information
Program Schedule
Table of Contents
Computer Networks, Architecture & High Performance Computing
Multicore Computation of Tactical Integration System in the Maritime Patrol Aircraft using Intel Threading Building Block
Muhammad Faris Falhoni, Bambang Sridadi
lmmersive Virtual 3D Environment based on 499 fps Hand Gesture Interface
Muhammad Sakli Alvissa/im
Improve fault tolerance in cell-based evolve hardware architecture
Chanin Wongyai
A New Patients' Rights Oriented Model of EMR Access Security
YB Dwi Selian/o, Yuslina Reino W Utami
Element Extraction and Evaluation of Packaging Design using Computational Kansei
iii
v
IX
x
xiii
7
13
19
Engineering Approach 25
Tazifik Djatna, Fajar Munichpulran/o, Nina Hairiyah, Elfira Febriani
Integrated Information System Specification to Support SSTC
Ahmad Tamimi Fadhilah, Yudho Giri Sucahyo, Denny
A Real Time Simulation Model Of Production System Of Glycerol Ester With Self Optimization
!wan Aang Soenandi. Taz1fik Djatna
33
39
Towards Maturity Model for E-Government Implementation Based on Success Factors 105
Darmawan Baginda Napilupulu
The Critical Success Factors to Develop an Integrated Application of Tuna Fishing Data Management in Indonesia
Devi Fitrianah, Nursidik Heru Praptono, Achmad Nizar Hidayanto, Aniati Murni Arymurthy
A Conceptual Paper on JCT as National Strategic Resources toward National Competitiveness} {Basuki Yusuf Iskandar and Fadhilah Mathar
Basuki Yusuf Iskandar and Fadhilah Mathar
Enterprise Computing
Quality Evaluation of Airline's E-Commerce Website, A Case Study of AirAsia and Lion Air Websites
Farah Shajira Effendi, lka Aljina
Hotspot Clustering Using DBSCAN Algorithm and Shiny Web Framework
Karlina Khiyarin Nisa
Analysis of Trust Presence Within Commerce Websites: A Study of Indonesian E-Commerce Websites
Muhammad Rijki Shihab, Sri Wahyuni, Ahmad Nizar Hidayanlo
The Impact of Customer Knowledge Acquisition to Knowledge Management Benefits: A Case Study in Indonesian Banking and Insurance Industries
Muhammad Rijki Shihab, Ajeng Anugrah Lestari
111
117
123
127
131
137
A System Analysis and Design for Sorghum Based Nano-Composite Film Production 143
Belladini Lovely, Tmifik Djatna
Moving Towards PC! DSS 3.0 Compliance: A Case Study of Credit Card Data Security Audit in an Online Payment Company
Muhammad R!fki Shihab, Febriana Misdianti
149
Social Network Analysis for People with Systemic Lupus Erythematosus using R4
Framework 217
Arin Karlina and Firman Ardiansyah
Experiences Using Z2SAL 223
Maria Ulfah Siregar, John Derrick, Siobhan North, and Anthony JH. Simons
Information Retrieval
Relative Density Estimation using Self-Organizing Maps
231
Denny
Creating Bahasa Indonesian - Javanese Parallel Corpora Using Wikipedia Articles 237
Bayu Distiawan Trisedya
Classification of Campus E-Complaint Documents using Directed Acyclic Graph
Multi-Class SVM Based on Analytic Hierarchy Process 245
Imam Cholissodin, Maya Kurniawati, lndriati, and Issa Arwani
Framework Model of Sustainable Supply Chain Risk for Dairy Agroindustry Based on
Knowledge Base 253
Winnie Septiani, Marimin, Yeni Herdiyeni, and Liesbetini Haditjaroko
Physicians' Involvement in Social Media on Dissemination of Health Information 259
Pauzi Ibrahim Nainggolan
A Spatial Decision Tree based on Topological Relationships for Classifying Hotspot Occurences in Bengkalis Riau Indonesia
Yaumil Miss Khoiriyah. and !mas Sukaesih Sitanggang
Shallow Parsing Natural Language Processing Implementation for Intelligent Automatic Customer Service System
Ahmad Eries Antares. Adhi Kusnadi, and Ni Made Satvika lnl'ari
SMOTE-Out, SMOTE-Cosine, and Selected-SMOTE: An Enhancement Strategy to Handle Imbalance in Data Level
265
27I
277
Application of Decision Tree Classifier for.Single Nucleotide Polymorphism Discovery
from Next-Generation Sequencing Data 335
Muhammad Abrar lsliadi, Wisnu Anania Kusuma, and I Made Tasma
v·
Alternative Feature Extraction from Digitized Images of Dye Solutions as a Model for
Algal Bloom Remote Sensing 341
Roger Luis Uy, Joel llao, Eric Punzalan, and Prone Mariel Ong
Iris Localization using Gradient Magnitude and Fourier Descriptor
Slewarl Sentanoe, Ania
S
Nugroho, Reggio N Harlono, Mohammad Uliniansyah,and Meidy Layooari
Multiscale Fractal Dimension Modelling on Leaf Venation Topology Pattern of Indonesian Medicinal Plants
Aziz Rahmad, Yeni Herdiyeni, Agus Buono, and Slephane Douady
347
353
Fuzzy C-Means for Deforestration Identification Based on Remote Sensing Image 359
Selia Darmawan Afandi, Yeni Herdiyeni, and Lilik B Prasetyo
Quantitative Evaluation for Simple Segmentation SVM in Landscape Image 365
Endang Purnama Giri and Aniali Murni A1y111urthy
Identification of Single Nucleotide Polymorphism using Support Vector Machine on
Imbalanced Data 37I
Lai/an Sahrina Hasibuan
Development of Interaction and Orientation Method in Welding Simulator for Welding Training Using Augmeneted Reality
Ario Sunar Baskoro, Aiohammad Azwar Amat, and Randy Pangestu Kuswana
Tracking Efficiency Measurement of Dynamic Models on Geometric Particle Filter
377
using KLD-Resampling 38 I
Alexander AS Gunm1•an. Wisnu Jatmiko. Vektor Dewan/a, F. Rachmadi, and F.
Jovan
Nonlinearly Weighted Multiple Kernel Learning for Time Series Forecasting
Agus Widodo. Indra Budi. and Be/awati Wicfiaia
385
'
ICACSIS 201-1
M Anwar Ma'sum, Wisnu Jatmiko, M Iqbal Tmi•akal, and Faris Al Afif
Particle Swarm Optimation based 2-Dimensional Randomized Hough Transform for
Fetal Head Biometry Detection and Approximation in Ultrasound Imaging 463
Putu Sah!'ika, Ikhsanul Habibie, M Anwar Ma'sum, Andreas Febrian, Enrico Budianto
Digital Library & Distance Learning
Gamified E-Learning Model Based on Community of Inquiry
Andika Yudha Utomo, Afifa Amriani, A/ham Fikri Aji, Fat in Rohmah Nur Wahidah, and Kasiyah M Junus
Knowledge Management System Development with Evaluation Method in Lesson Study Activity
Murein Miksa Mardhia, Armein ZR. Langi, and Yoanes Bandung
Designing Minahasa Toulour 3D Animation Movie as Part of Indonesian e-Cultural Heritage and Natural History
Stanley Karouw
Learning Content Personalization Based on Triple-Factor Learning Type Approach in
E-learning
Mira SwJ1ani, Hany Budi Sanloso. and Zainal A. Hasibuan
Adaptation of Composite E-Learning Contents for Reusable in Smartphone Based Learning System
Herman Tolle, Kohei Arai. and Alyo Pinandito
The Case Study of Using GIS as Instrument for Preserving Javanese Culture in a
469
477
483
489
497
Traditional Coastal Batik, Indonesia 503
TJi beng .lap and Sri Tiatri
Identification of Single Nucleotide Polymorphism
using Support Vector Machine on Imbalanced Data
Lailan Sahrina Hasibuan, Wisnu Anania Kusuma, Willy Bayuardi Suwarno
Deparrmenl ofCompuler Science, Faculry of Malhema/ics and Narura/ Sciences, Bogar Agriculrural Universiry
Deparlmenl of Agronomy and Horriculrure, Faculry of Agriculture, Bogar Agriculrural Universiry
E-mail: [email protected], [email protected], [email protected]
Abstract-The advance of DNA sequencing tech110/ogy presents a sig11iflca11t bioi11for111atic
chal/e11ges in a do11111strea111 analysis such as
identification of single nucleotide poly111orphis111 (SNP), SNP is the most ab1111da11t form of genetic
111arker and hai•e been one of the 1110s/ crucial
researches in bioinfor111alics. SNP has been applied
in wide area, but analysis of SNP in plants is very• limited, as in cu//ivated soybean (Glycine ャャエャャセ@ l.).
This paper discusse; the identijicatio11 of SNP i11
culfil'aled soybean u.-;ing Support Vector Machine
(SVM). SVM is trained using positfre and negative
SNP. Previous/)1, we perfor111ed a balancing positive
and negative SNP with u11dersa111pling and
oversa111pling to obtain training <lata. As a result, the 111otlel 1vhich is trainetl 1vitfl balancetl tlatll has better perfor111a11ce tlta11 that with i111bala11ced tlllta.
Keywords- identitkalion; SNP; SVM;
oversampling; undcrsampling.
I. INTRODUCTION
T
HE latest technological advancement in DNAsequencing. is the next generation of sequencing
(NOS). NOS is also referred as high-throughput DNA sequencing (HTS). NOS or HTS has reduced the cost and increased the throughput of DNA sequencing because its ability to automate techniques in DNA sequencing [I]. However, NOS has shortcomings in the quality of the sequencing results, it produce shorter read lengths than previous technology, Sanger method [2]. It has a tremendous impact on how the reads have
to be processed in a do\vnstrean1 analysis such as
identification of Single Nucleotide Polymorphism (SNP).
SNP is the most abundant forn1 of genetic 111arker in
セᄋャ。ョオウ」イゥーエ@ rccci\·cd July 28. 201-1.
lailan S. H and \\'. A. Kusuma arc with 1hc Department of
Con1putcr Science . Faculty of セャ。エィ」ュ。エゥ」ウ@ and Natural
Sciences. Bogar 1\griculiural University. Bogar 16680 H」セュ。ゥャZ@
lailan.sahrina.hsb'il gmail.com. ananta'll ipb.ac.id).
\\'. B. sオキ。セQP@ is with d」ー。セュ」ョエ@ of Agronon1y and
llorticulturc. Faculty of Agricul!urc. Rogor Agricultural Univcrsily.
Bogor 16680 (e-mail: hayuardi'ci gmail.com ).
371
all populations of individuals [3]. SNP is defined as
single base variations or short insertions or deletions in the nucleotide sequence from different individuals or between homologous sequences within an
individual [4].
NOS technologies for SNP analysis has been applied in wide area such as medicine [5], animal genetics [6] and plant breeding [7]. However, SNP analysis in plants such as cultivated soybean (Glycine
max l.) is limited [3].
This paper will focus on identification of SNP in cultivated soybean using Support Vector Machine (SVM). SVM is an effective method for general
purpose supervised pattern recognition and has been
applied successfully to many biological problems
recently, such as gene selection [8], cancer
classification [9] and detection of cardiac abnormality
[I OJ.
This paper is organized as follows. Section II will discuss about research related to identification of SNP that have been done before. Section Ill presents techniques at data level for pre-process, features used to train SVM, techniques to handling imbalance of
data, undersampling and oversampling, training and testing \vith SVM. Results, discussion and conclusions
are presented in section IV, V and VI.
II. RELATED WORK
Research about identification of SNP have been done both ·an DNA of human, animals and plants. In [4] they developed a machine learning to identify SNP on DNA of cultivated soybean. They used decission
tree as classifier and feed fonvard neural net\vork as
learning method. The training data were 27.275 candidate SNP generated by sequencing 1973 STS (Sequence Tag Sites) from 6 diverse homozygous
soybean cultivar. The accuracy reaches 97 .3% \\'ith
PPV of 84.8%.
Kong and Choo developed a method based on SVM
(Support v・」エッイセm。」ィゥョ・I@ to predict SNP. The
research was conducted on human DNA. One-thousand positive SNP and I 000 negative SNP were randomly selected from JSNP, SNP database of the
ICACSIS 2014
Sampling outcome data, undersampling and
oversampling. were divided into k subsets (k = I 0). Of
the k subsets, a single subset was retained as the testing data, and the remaining k - 1 subsets \Vere used as training data. The cross-validation process is then
repeated k times, with each of the k subsets used exactly once as the testing data. We did undersampling process two times with a value of m
= I
and m=
2.We also conducted experiments on imbalanced data. We divided the data into k subsets (k = I 0). Of the k
subsets, a single subset was retained as the testing
data, and the remaining k - I subsets were used as
training data .. The cross-validation process is then
repeated k times, \vith each of the k subsets used
exactly once as the testing data.
IV. RESULT
Identification of SNP that we did are divided into four categories: (I} identification of SNP using undersampling data with m
=
I, (2) m=
2, (3) oversampling data, and (4) imbalance data. We usedmetrics based on confusion matrix as sho\vn in Figure 1 for evaluation. Confusion matrix has four categories:
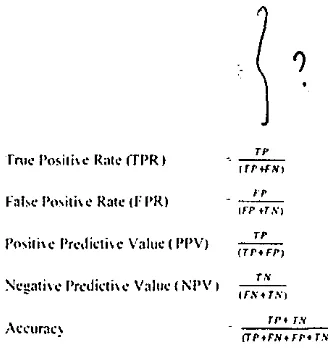
True positives (TP) are examples correctly labeled as positives. False positives (FP) refer to negative examples incorrectly labeled as positive. True negatives (TN) correspond to negatives correctly labeled as negative. Finally., False negatives (FN) refer to positive examples incorrectly labeled as negative [21 ].
We plotted the FPR (False Positive Rate) and TPR (True Positive Rate) of each categories (Figure 2). From this plotted we can see that category (I), (2), and (3) have better performance than category (4) because they have higher TPR and lower FPR. Generally, when TPR is high, then FPR is also high. This condition n1eans that \vhen a classifier has high ability
in indentifying positive SNP as positive SNP then this classifier also has high fault in identifying negative
SNP as positive SNP. The two criteria, TPR and FPR,
are エイ。、・セッエtN@ \\'e cannot use one of then1 to evaluate performance of a classifier. Hence \\'e need a metric
that combine the ability of classifier in indentifying positive SNP as positive SNP and the ability of
I l'n:JkliC1n
I rarccl I T ! f
! r ' TP I FN
'
L f I FP
I
TN0
,.
00 セ@
0
1' セ@ :;
"
O'. 0
'
セ@ !
0
-Q_
•
.,
,.f'.
0 '; I : セ@
セ@ f
N
-"
0 !J
!; • m::l
,•
0 I
0
• m::2
• <MBamphng
• imbalillltt
00 02 04 06 08 1 0
False Positive Rale
Fig I. Plotted of false potive rate and true positive rate. Black
dots shows the results of identification using training data undersampled with m =I, blue dots ism= 2, green dots is
oversampling, and red dots is imbalance data
classifier to not identify negative SNP as positive
SNP. We used Fmeasure to measure this metric,
precision is refer to PPV and recal is refer to TPR.
2 • Precision • Recall
Precision+ Recall (2)
We also measured G-Mean, the geometric mean of
classifier performance on two classes separately.
TP s1ms it iv it)'
=
_T_P_+_F_N_TN
specificity= TN+ FP (3)
G - .'ifean
=
,/ssnsitiftty "speciffcityFrom each category, \Ve choosed the best classifier based on Frneasure· The best classifier of each category
is presented in Table II. FPR and TPR plotted of best
classifier, is presented in Figure 3.
rnu: pッセゥQィ」Z@ Ra!!.• (fPR)
"
rrr+FN!
,,
lfl' tT,\')
"
(TftFJ')
"'
4f.\t TS)rr+ r.v
Fig 2. Co1nmon nmchinc Jcan1ing cvalm11ion metrics
[image:15.603.304.508.76.260.2] [image:15.603.288.452.579.754.2]セャ・。ョ@ base quality (#9): mean quality of all bases at the \"ariant
posi1ion.
Alignment depth (#10): number of reads aligned at variant
position. セ@
Error probability (# 11 ): probability of the number of the reads containing -\"ariant base was sampled from binomial distribution
with gi\'en parameters.
Dinucleotide repeat (#12.13): number of dinucleotide repeat around the variant position (left and right direction).
セャゥウュ。エ」ィᄋ。イ・。@ (#1-t): mean number of variant base per each reads aligned a1 \'ll!iant position.
Homopolymer length (#15.16): length of repeating bases around !he variant position {left and right direction).
!"ucleotid; diversity(# 17): deviation of reference base frequencies from given whole-genome average.
Total mismatch count (#18,19): number of mismatch on reads
with reference base and reads with variant base.
Allele balance (#20): number of reads containing variant bases divided by セfァョュ・ョエ@ depth.
セャ・。ョ@ of Nセ・。イ「ケ@ base quality (#21): mean quality of all bases around variant position.
Distance エセ@ nearest ,·ariant (#22,23): distance of the variant to its ョ・ゥァィ「ッイゥョセ@ variant (left and right flank size).
ACKNOWLEDl}MENT
The authors would like to thank to Abrar lstiadi,
a
member ... of the Laboratory of Bioinformatics, Depanme.nt of Computer Science, Boger Agricultural Universify (!PB), for his very helpful assistance on the extraction of SNP features.JI) )2) (31 )4) [5) )6) REFERENCES
M. L Metzker. "Sequencing technologies - the ョセクエ@
generation:· i\'al. Rev. Genet .. vol. 11. no. I. pp. 31--46. Jan. 2010.
Elaine R. Mardis, "The impact of next-generation sequencing technology on genetics:· Trends Genet. ,·ol. 24. no. 3. pp 133· 141. Feb. 2008
S. Kumar, T. \\I. Banks, S. Cloutier. "SNP discovery through Next-Generation Sequencing and its applications.·· /111 J Plant
Ge11omics. vol 2012. Nov. 2012
L. K. tdatukumalli. J. J. Grefenslette. D. L. Hytcn.1.-Y. Choi. P. B .. ·Cregan. and C. P. Van Tassell. "Application ofnrnchine learning セ@ SNP discovery:· B,\IC Bioinformatics. vol. 7. no. 4. Jan. 2006
Nikolina BahiC. "Clinical pharmacogcnomics and concept of personalized medicine.'' J ,\ft:d Biocl11:m. vol. 31. no.4. pp 281-6.
A. Vi2nal. D tvfilan. tvl. S. Cristobal. A. Eggen. "A review on SNP ;nd other types of molecular n1arkers and their use in animal genetics:· Genet. Se/. El·ol, vol. 34. no. 3. pp. 275-305. Jun 2002
[7] J. lvlammado\', R. Aggan\'al, R. Bu))'arapu, and S. Kumpatla, '"SNP markers and their impact on plant breeding." Int. J.
Plant Genomics, vol. 2012, Jan. 2012.
[8] X. Zhou and D. P. Tuck, "MSVM-RFE: extensions of
SVtvf-RFE for multiclass gene selection on DNA microarray data ...
Bioinformaticsl. vol. 23, no. 9, pp. 1106-14, Sep. 2007,
{9] A. Bharathi and A.M. Nataffijan. ··cancer classification using support \'CCtor machines and relevance vector machine based on analysis of variance features:· J. Con1p111er Sci, vol. 7, no. 9, pp. 1393-9, Sep. 201 L
[10] S. Ari, K. Hembram, G. Saha, "Detection of cardiac abnormality from PCG signal using LMS based least square SVM classifier." Expert Syst Appl, vol. 37, pp. 8019-26, Dec. 2010.
[I I] \\'. Kong and K. \\'. Choo. "Predicting single nucleotide pol)morphisms (SNP) from DNA sequence by support vector machine:· Front,Biosci, pp. 1610-4, Jan. 2007.
[12] 8. D. O'Fallon. W. Wooderchak-Donahue, and O. K.
Crockett. ··A support vector machine for identification of single-nucleotide polymorphisms from next-generation sequencing data:' Bioinjormatics, vol. 29, no. 11, pp. 1361-6, Jun. 2013.
[13] H. Xiong, et.al, ··A hybrid Fisher/SVM method for SNP discovery in Brassica oilseed rape." J Food Agr Environ,
vol.8, no. 3&4, pp. 705-8, Okt. 2010.
[14] H.M. Lam, el al., "Resequencing of 31 wild and cultivated soybean genomes identifies patterns or genetic diversity and selection." Nat. Genet., vol. 42. no. iセN@ pp. 1053-9, Dec. 2010.
[ 15] J. Schmutz, el al .. "Genome sequence of the palaeopolyploid soybean.," Nalure, vol. 463, no. 7278. pp. 178-83, Jan. 2010. [ 16 J R. Li, el al., "SOAP2: an improved ultrafast tool for short read alignment:· Bioinformatics, vol. 25, no. 15, pp. 1966-7. Jun 2009.
(17] J. Van Oeveren and A. Janssen. "Mining SNPs from DNA Sequence Data: Computational Approaches to SNP Discovery and Analysis," in Single Nucleotide Po(vmo1phisms, vol. 578, A. A. Komar, Ed. Totowa, NJ: Humana Press, 2009, pp. 73-91.
[18] S. J. Yen, Y. S. Lee. "Cluster-based under-sampling approaches for in1balanccd data distributions." £.\per/ Syst
Appl. vol. 36. pp. 5718-27. Apr. 2009.
( 19) R. Batuwita. and V. Pa lade. "Efficient rcsampling methods for training support vector machines. with imbalanced datasets." In 1\'e11ra/ i\'etworks (IJCNX). The 2010
l11ter11atio11al Joint Conference 011. pp. 1-8. IEEE. 2010.
[20] D. Meyer. et al. c I 071: Misc Functions of the Department of Statistics (e1071 ). TU Wien. R package version 1.6-3. http://CRAN.R-project.org/package==el 071. 2014.
[21) Y. Tang. et.al. "SVt-.1s modeling for highly imbalanced classification." Systems . . \fan. a11d Cyber11et1cs. Part B:
Cybernelics. /£££Transactions vol. 39. no. I. pp. 281-288.
Jan. 2009.
[22] R. Batuwita. and V. Palade. "Class imbalance learning methods for support vector machines"' In: He. H .. Ma. Y. (Eds.). Imbalanced Leaming: Foundations. Algorithms. and 1\pplications. in press. \Viley. 2013