COMPARATIVE STUDY OF ENDOPHYTIC BACTERIAL
COMMUNITY STRUCTURES IN FOUR INDONESIAN
RICE CULTIVARS BASED ON 16S rRNA SEQUENCE
YENI KHAIRINA
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
THE THESIS STATEMENT AND SOURCES OF INFORMATION ALONG COPYRIGHT DEVOLUTION
I hereby declare that the thesis entitled “Comparative study of
endophytic bacterial community structures in four Indonesian rice cultivars based on 16S rRNA sequence” is true of my research under the guidance of the supervisor committee and has not been submitted in any form to any college. Sources of information derived or citated from published and unpublished works from other writers have been mentioned in the text and listed in the references in the end of this thesis.
I hereby assign the copyright of my thesis to Bogor Agricultural University.
Bogor, March 2016
Yeni Khairina
Empat Kultivar Padi asal Indonesia berdasarkan Sekuen 16S rRNA. Dibimbing oleh YULIN LESTARI dan ANJA MERYANDINI.
Mikrob endofit telah banyak dilaporkan memiliki pengaruh yang positif bagi tanaman inangnya, misalnya dengan mendukung pertumbuhan tanaman, memperkuat pertahanan melawan bakteri patogen, dan meningkatkan asupan nutrisi. Namun, studi tentang diversitas mikrob endofit khususnya pada kultivar padi asal Indonesia dengan agro-ekosistem yang berbeda masih terbatas disebabkan banyaknya penggunaan metode kultur biasa. Denaturing Gradient Gel Electrophoresis (DGGE) merupakan salah satu pendekatan metagenomik berdasarkan pada pemisahan fragmen-fragmen DNA dengan ukuran yang sama namun memiliki sekuen basa yang berbeda. Metode ini dapat mengungkapkan diversitas mikrob yang sulit untuk dikulturkan dalam media buatan. Berdasarkan latar belakang di atas, penelitian ini bertujuan untuk menganalisis dan membandingkan struktur komunitas bakteri endofit dan spesifik takson pada bakteri yaitu aktinomiset, menggunakan gen 16S rRNA pada empat kultivar padi asal Indonesia dengan metode PCR-DGGE.
Bagian tanaman (akar, batang, dan daun) yang sehat diambil dari 4 kultivar padi asal Indonesia yaitu Ciherang, IR64, Inpara 2, dan Situ Patenggang, DNA total diekstraksi menggunakan Genomic DNA Mini Kit Plant. Amplifikasi PCR untuk komunitas bakteri dilakukan pada sampel akar, batang, dan daun tanaman padi yang menghasilkan panjang fragmen sekitar ±600 bp. Sementara itu, amplifikasi PCR untuk komunitas aktinomiset dilakukan pada sampel batang dan daun menggunakan strategi PCR 2 tahap yang menghasilkan fragmen dengan ukuran sekitar ±1087 bp (PCR pertama) dan ±195 bp (PCR kedua). Analisis diversitas untuk komunitas bakteri dan aktinomiset endofit dilakukan dengan metode DGGE pada gel poliakrilamid. Pita target dipotong dan kemudian diamplifikasi kembali dengan menggunakan primer tanpa GC clamp. Sekuensing pada produk PCR dari pita DGGE dilakukan sesuai dengan standar protocol menggunakan ABI PRISM sequencer. Analisis hubungan antar DNA yang disekuensing dilakukan berdasarkan pohon filogeni menggunakan metode
neighbor-joining pada perangkat lunak MEGA 5.0.
Analisis Shannon-Wiener dan profil DGGE menunjukkan diversitas bakteri endofit pada padi kultivar Ciherang dan IR64 lebih tinggi dibandingkan dengan Inpara 2 dan Situ Patenggang. Analisis dice similarity coefficient
2601, and Pseudomonas brassicacearum subsp. brassicacearum NFM421 strain NFM421 dengan identitas kesamaan berkisar antara 90-97%. Kelas β -Proteobacteria memiliki afiliasi spesies seperti Burkholderia cepacia strain 106
dengan identitas kesamaan 93%. Kelas α-Proteobacteria memiliki afiliasi spesies
yaitu Brevundimonas olei strain DUCC3718 dengan identitas maksimum 92%. Kelas Bacilli memiliki afiliasi spesies seperti Sporosarcina koreensis strain F73 dan Brevibacillus brevis DZBY12 dengan identitas kesamaan masing-masing 97% dan 99%. Kelas Flavobacteria memiliki afiliasi spesies seperti Myroides odoratus
dan Flavobacterium ceti strain 454 dengan identitas kesamaan 91-99%. Sementara itu, satu pita teridentifikasi sebagai archaea.
Hasil analisis Shannon-wiener dan profil DGGE pada spesifik takson bakteri, aktinomiset, menunjukkan nilai diversitas dari komunitas ini tidak terlalu berbeda antar sampel yang dibandingkan. Analisis dice similarity coefficient dan analisis kluster menunjukkan adanya kesamaan struktur komunitas yang tinggi antara padi kultivar Ciherang dan IR64. Analisis filogeni menunjukkan kluster aktinomiset dengan 4 famili besar yaitu Microbacteriaceae, Streptomycetaceae, Cellulomonadaceae, dan Micrococcaceae. Famili Microbacteriaceae memiliki afiliasi spesies seperti Microbacterium insulae strain DS-66 dan Microbacterium luteolum strain IFO 15 074 dengan identitas maksimum 99%. Famili Streptomycetaceae memiliki afiliasi spesies seperti Streptomyces acidiscabies
strain ATCC 49003, Streptomyces chartreusis strain ISP 5085, dan Streptomyces scopiformis strain NBRC 100 244 dengan identitas maksimum 98-99%. Famili Cellulomonadacee memiliki afiliasi spesies yaitu Cellulomonas flavigena strain DSM 20109 dengan identitas maksimum 100%. Famili Micrococaceae memiliki afiliasi spesies seperti Kocuria polaris strain CMS 76or, Kocuria rosea strain DSM 20447, Kocuria aegyptia strain YIM 70003, Arthrobacter aurescence TCI strain TCI, Arthrobacter ramosus strain CCM 1646, Arthrobacter arilaitensis
strain Re117, Citricoccus nitrophenolicus strain PNP1, dan Micrococcus luteus
NCTC strain 2665 dengan identitas maksmum 99-100%.
YENI KHAIRINA. Comparative Study of Endophytic Bacterial Community Structures in Four Indonesian Rice Cultivars based on 16S rRNA Sequence.
Supervised by YULIN LESTARI and ANJA MERYANDINI.
Endophytic microbes have been reported to give beneficial effects to their host by promoting the plant growth, strengthening the protection against pathogen, and increasing nutritional supply. However, studies on the diversity of microbial endophytes especially in Indonesian rice cultivars with different agro-ecosystems are still limited due to the mainly use of culture-dependent method. DGGE (Denaturing Gradient Gel Electrophoresis) is one of metagenomic approach based on the separation of the DNA fragments that have the same length but with different sequences. It can be used to reveal the diversity of microorganisms that are difficult to be cultured in the artificial media. Thus, this study aimed to compare the community structure of bacterial endophytes and specific bacterial taxon, actinomycetes, based on 16S rRNA gene in four Indonesian rice cultivars using PCR-DGGE method.
The part of plant samples (root, stem, and leaf) were collected from four healthy rice cultivars IR 64, Inpara 2, Situ Patenggang, and Ciherang. Total DNA was extracted using Genomic DNA Mini Kit Plant. PCR amplification of bacteria domain was done from root, stem, and leaf samples resulted in fragment size ±600 bp. Meanwhile, PCR amplification of actinomycetes was done from leaf and stem using two-stage PCR strategy resulted fragment size ±1087 bp (first PCR) and ±195 bp (second PCR). Diversity analysis of endophytic bacteria and actinomycetes community was conducted using DGGE on polyacrylamide gel. Bands of interest were excised and re-amplified using the primer without GC-clamp. Sequencing of re-amplification product of DGGE bands was done according to standard protocols using DNA sequencer ABI PRISM. The sequencing results were compared to the GenBank nucleotide sequence database of NCBI BLAST.N. Relationship analysis among the sequenced DNA was performed based on phylogenetic tree using the neighbor-joining method and software MEGA 5.0
Shannon-Wiener analysis and DGGE profiles showed the diversity of endophytic bacteria in Ciherang and IR64 rice cultivars were higher compared with that of Inpara 2 and Situ Patenggang. The dice similarity coefficient showed that all of the samples have a quite similar community structures, however cluster analysis of endophytic bacteria demonstrated high similarity of the community structure between Ciherang and IR64 rice cultivars and community structure between Inpara 2 and Situ Patenggang. Distributions of bacterial members to phylogenetic taxon were varied among the rice cultivars in which the majority of the sequences obtained were closely related to Gammaproteobacteria, Bacilli,
Flavobacteria, β-Proteobacteria, and α-Proteobacteria. Gammaproteobacteria class
was affiliated to Cellvibrio japonicus strain UEDA 107, Pseudomonas putida
strain ZJUTBX04, Escherichia fergusonii strain NBRC, Pseudomonas poae
RE*1-1-14 strain RE*1-1-14, Cellvibrio mixtus strain J3-8, Cellvibrio mixtus
NFM421 strain NFM421 with 90-97% of maximum identity. β-Proteobacteria
class was affiliated to Burkholderia cepacia strain 106 with 93% of maximum
identity. α-Proteobacteria was affiliated to Brevundimonas olei strain DUCC3718
with 92% of maximum identity. Bacilli class was affiliated to Sporosarcina koreensis strain F73 and Brevibacillus brevis DZBY12 with 97% and 99% of maximum identity, respectively. Flavobacteria class was affiliated to Myroides odoratus and Flavobacterium ceti strain 454 with 91-99% of maximum identity. Meanwhile, one band was identified into archaea.
Focusing on specific bacterial taxon, actinomycetes, Shannon-Wiener analysis and DGGE profiles showed the diversity of this community were not different among the samples. The dice similarity coefficient and cluster analysis of endophytic actinomycetes demonstrated high similarity of the community structure between Ciherang and IR64 rice cultivars. Phylogenetic analysis showed actinomycetes cluster with 4 large families, they were Microbacteriaceae, Streptomycetaceae, Cellulomonadaceae, and Micrococcaceae. Microbacteriaceae was affiliated to Microbacterium insulae strain DS-66 and Microbacterium luteolum strain IFO 15 074 with maximum identity 99%. Streptomycetaceae was affiliated with Streptomyces acidiscabies strain ATCC 49003, Streptomyces chartreusis strain ISP 5085 and Streptomyces scopiformis strain NBRC 100 244 by maximum identity 98-99%. Cellulomonadaceae was affiliated with
Cellulomonas flavigena strain DSM 20109 with maximum identity 100%. Micrococaceae was affiliated to Kocuria polaris strain CMS 76or, Kocuria rosea
strain DSM 20447, Kocuria aegyptia strain YIM 70003, Arthrobacter aurescence
TCI strain TCI, Arthrobacter ramosus strain CCM 1646, Arthrobacter arilaitensis
strain Re117, Citricoccus nitrophenolicus strain PNP1, and Micrococcus luteus
NCTC strain 2665 with maximum identity 99-100%.
© Copyright of IPB, 2016 Copyright Reserved
Prohibited for quoting part or all of this thesis without including or citing the sources. Citation is only for educational purposes, research, scientific writing, report writing, criticism writing, or review of an issue; and citations are not detrimental on behalf to IPB.
COMPARATIVE STUDY OF ENDOPHYTIC BACTERIAL
COMMUNITY STRUCTURES IN FOUR INDONESIAN
RICE CULTIVARS BASED ON 16S rRNA SEQUENCE
YENI KHAIRINA
Thesis
as one of the requirements to obtain the degree Master of Science
on
Microbiology Major
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
Thesis Title : Comparative Study of Endophytic Bacterial Community Structures in Four Indonesian Rice Cultivars based on 16S rRNA Sequence
Name : Yeni Khairina
NIM : G351124121
Approved by Supervisor Commission
Dr Yulin Lestari Prof Anja Meryandini, MS
Head Member
Discovered by
Head of Microbiology Dean of Graduate School
Major
Prof Dr Anja Meryandini, MS Dr Ir Dahrul Syah, MScAgr
completed successfully. The theme chosen in this research was endophytic microbial diversity that has been conducted from July 2014 until March 2015. The
title of this thesis research is “Comparative Study of Endophytic Bacterial
Community Structures in Four Indonesian Rice Cultivars Based on 16S rRNA Sequence”.
The author thanks to Dr.Yulin Lestari and Prof. Dr. Anja Meryandini MS.as supervisor.Thanks is also delivered to Prof. Yasuyuki Hashidoko that has supervised the author during Population Activities Resources and Environment (PARE) exchange program. The author also thanks to Dr. Ir. I Made Artika, M.App.Sc. as an external examiner has given advices during thesis examination. The author also thanks to the staff of Laboratory of Microbiology and Intergrated Laboratory, Department of Biology IPB, and Laboratory of Ecological Chemistry, Hokaido University.
During the study and research, the author thanks to the author’s family. The author also gives special thanks to Mahyarudin, Randi, Sari, Mei, Reika, Sharon, Nie, Ciko, Sipri, Gegek, Chessa, Septi, Sasha, Rara, Wahyu, Vita, Asril, all of friends in Microbiology Major Batch 2012-2013, Graduate School of IPB, all of friends in Laboratory of Ecological Chemistry, and all of PARE members who have helped the author during this research. The author wished this research can give contribution for the knowledge development specifically in agricultural sector.
Bogor, March 2016
TABLE OF CONTENTS
LIST OF TABLES xiv
LIST OF APPENDIX xv
INTRODUCTION 1
Background 1
Problem Identification 2
Objective of Study 2
Significant of Study 2
Research Scope 3
LITERATURE REVIEW 3
Endophytic Bacteria 3
Rice Plant 7
PCR-DGGE 8
METHODS 9
Research Framework 9
Time and Place 10
Sample Collection and Sterilization 10
DNA Isolation 11
Amplification of 16S rRNA Gene-specific Bacteria and Actinomycetes 12
Analysis and Cloning of DGGE Bands 13
Sequencing of the 16S rRNA Gene and Construction of Phylogenetic Tree 14
Sequence Variation Analysis 15
RESULT AND DISCUSSION 15
Result 15
Discussion 31
CONCLUSION AND SUGGESTION 34
Conclusion 34
Suggestion 34
REFERENCES 36
APPENDIX 43
screened by SEM at 7 dpi 5
2 Research framework 10
3 PCR amplification of 16S rRNA gene specific bacteria from four
Indonesian rice plant cultivars 16
4 Bacterial community in four Indonesian rice cultivars 17
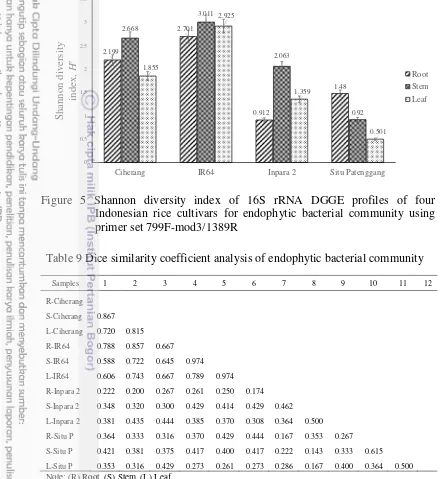
5 Shannon diversity index of 16S rRNA DGGE profiles of four Indonesian rice cultivars for endophytic bacterial community 18 6 Hierarchical cluster analysis results of bacteria DGGE profiles 19 7 The closest sequence homology of bacterial 16S rRNA gene-targeted
DGGE band 21
8 Position of DGGE band region amplified by 799F-mod3/1389R
primer set 22
9 PCR amplification of 16S rRNA gene specific actinomycetes from
four Indonesian rice plant cultivars 24
10 Actinomycetes community in four Indonesian rice cultivars 25
11 Shannon diversity index of 16S rRNA DGGE profiles of four Indonesian rice cultivars for endophytic actinomycetes community 26 12 Hierarchical cluster analysis result of actinomycetes DGGE profiles 26 13 The closest sequence homology of actinobacterial 16S rRNA
gene-targeted DGGE band 28
14 Position of the actinomycetes community amplified by 338F/518R
primer set 29
LIST OF TABLES
1 Endophyte classification based on colonization in the plant tissue 4
2 Endophyte classification based on the lifestyle 4
3 Rice plant profile in Indonesia 7
4 Some endophytic bacteria isolated from rice plant 8
5 Characteristics (I) of agricultural soil from sampling site 11
6 Characteristics (II) of agricultural soil from sampling site 11
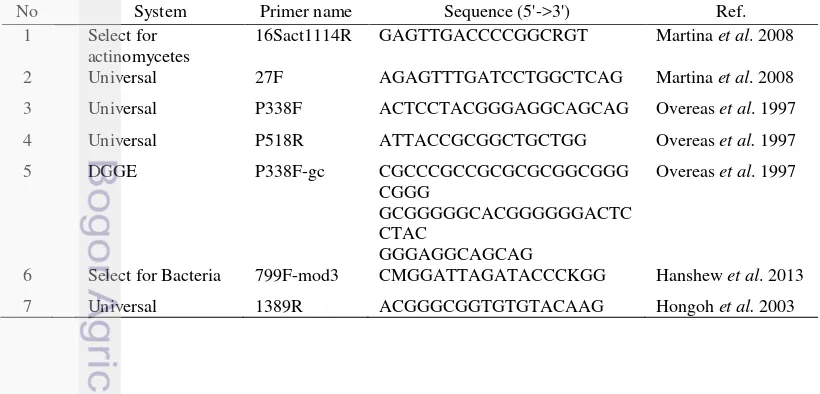
7 List of the primers used in this study 12
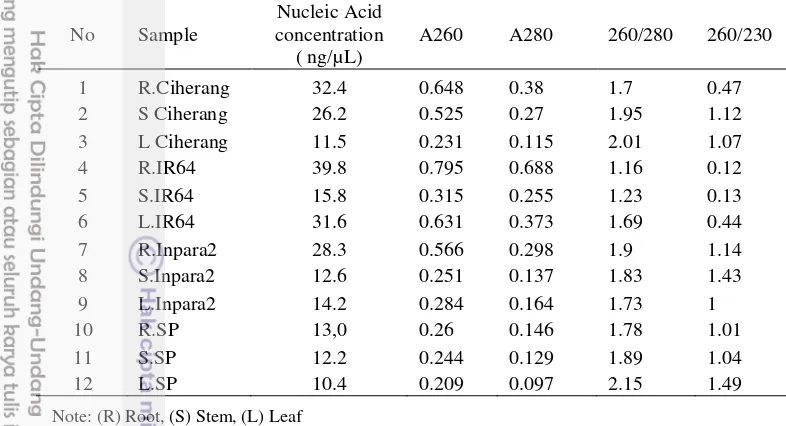
8 The quantity and quality of DNA extracted from rice plant tissue 16 9 Dice similarity coefficient analysis of endophytic bacterial community 18
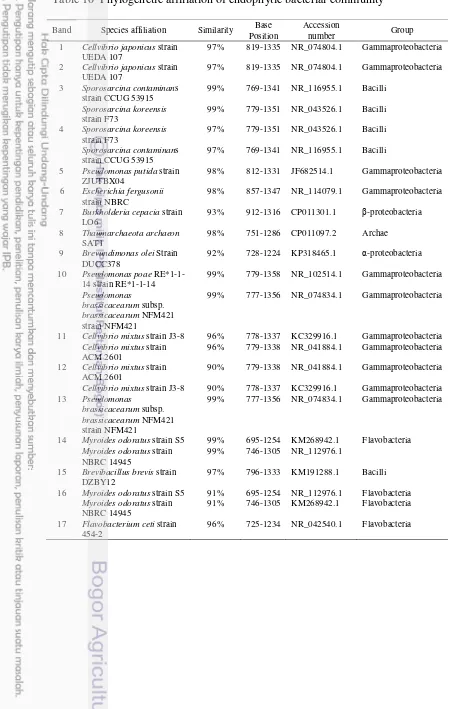
10 Phylogenetic Affiliation of Endophytic Bacterial Community 20
11 Variation of specific sequence pattern in DGGE bands originated from
Indonesian rice plant 23
12 Dice similarity coefficient analysis of endophytic actinomycetes
community 26
13 Phylogenetic Affiliation of Endophytic Actinomycetes Community 27
LIST OF APPENDIX
1 Sequence of DGGE bands for bacterial community based on 16S
rRNA gene 43
2 Sequence of DGGE bands of actinomycetes community based on 16S
rRNA 48
3 Variation pattern of DGGE bands for bacterial community 50
4 Data of DGGE band volume from Phoretix 1D software for bacterial
community 56
5 Data of DGGE bands volume from Phoretix 1D software for
actinomycetes community 57
INTRODUCTION
Background
Indonesia is the third-largest rice producer in the world (USDA 2015). However, the forecast data showed the increase of rice imports around 2% in 2015 compared than previous year (Statista 2015). It indicated that the high rate of rice production cannot achieve self-sufficiency of the greater demand of rice. High demand of rice can be attributed due to the population increase. In order to achieve food stability and security in Indonesia, various efforts should be conducted, one of them is by increasing rice productivity. The improvement of cultivation techniques by utilizing endophytic microbes is one of good alternatives to achieve both food and environment sustainability.
In recent years, much attention has been paid to the study of endophytic microbes due to their great potential as less exploited resources. Some of the endophytic microbes were reported to give beneficial effects to their host by promoting the plant growth, strengthening the protection against pathogen, and increasing nutritional supply(Singh et al. 2006; Nagendran et al. 2014; Sari et al.
2014). A wide range of bioactive compounds has also been produced by endophytic microbes, which were commonly derived from bacterial group especially a member of phylum Actinomycetes (Berdy 2005). These microbes have huge potential to synthesis numerous novel compounds that can be applied in pharmaceutical, agricultural and industries(Golinska et al. 2015). Nowadays, the exploration of microbial diversity of endophytes and their potential was still limited in the certain types of plants. Thus, the novel potential of endophytes and their distribution throughout the diverse plant species in various ecosystems is still being a subject of interest.
Practice in agriculture is one of the factors known to affect the diversity of microbial community in the soil (Lopes et al. 2011), but very little knowledge about the effects of these practices on the existence of endophytic microbial communities, especially in rice plant. Some of superior rice cultivars in Indonesia are very interesting to be studied, for example IR 64 and Ciherang that currently hold about 31% and 22% of total national rice area, respectively (USDA 2012), and the other varieties like Inpara 2 and Situ Patenggang that survive very well in unfavorable environmental condition. Those rice cultivars have been adapted to different kind of agro-ecosystems and cultivation techniques. IR 64 and Ciherang are commonly cultivated in irrigated rice field, Inpara 2 is planted on tidal swampland, while other rice variety such as Situ Patenggang is well adapted in dry land (Suprihatno et al. 2009).
known that only small portion of microbes (0.1-10%) from the total population can be culturad. Meanwhile, more than 99% of microbes are still difficult to culture in artificial media (Sekiguchi 2006). A solution to overcome the difficulties and limitations associated with the cultivation technique is metagenomic approaches.
DGGE (Denaturing Gradient Gel Electrophoresis) is one of a metagenomic approach based on the separation of the DNA fragments that have the same length but with different sequences (Fischer and Lerman 1994). The use of PCR-DGGE technique in the study of biodiversity has benefits to track and describe the dominant population within the samples. In its application, DGGE is applicable for overviewing the succession and diversity of microbial community structure because it can proceed many different samples simultaneously based on environmental change (Piterina et al. 2012). Using PCR-DGGE, Mahyarudin et al. (2015) has successfully revealed the community structure of actinomycetes in soil and root of some Indonesian rice plants, including four cultivars as described above. To obtain more comprehensive data, further DGGE analysis of actinomycetes community in stem and leaf as well as bacterial community structure in root, stem, and leaf of these Indonesian rice varieties were done. This study aimed to compare the community structure of bacterial endophytes and a specific bacterial taxon, Actinomycetes, in the Indonesian rice cultivars using 16S rRNA gene-targeted PCR-DGGE analysis.
Problem Identification
Endophytic bacteria have been reported to have an important role for supporting the growth of rice plants. However, data of the overall diversity of endophytic bacteria in Indonesian rice plants with difference agro-ecosystems are still not available yet. It is because only 1% of microbes that can be cultured, whereas approximately 99% of microorganisms cannot be cultured in artificial media.
Objective of Study
This study aimed to compare the community structure of bacterial endophytes and specific bacterial taxon, actinomycetes, based on 16S rRNA gene in four Indonesian rice cultivars using PCR-DGGE analysis.
Significant of Study
Research Scope
The research involves sample collection and sterilization, the isolation of genomic DNA in rice plants tissues, the amplification of 16S-rRNA gene-specific bacteria and actinomycetes, diversity analysis using PCR-DGGE, and phylogenetic tree construction.
LITERATURE REVIEW
Endophytic Bacteria
Endophyte is derived from "endon" which means "inside" and "python" which means plant (Schulz and Boyle 2006). Endophytism is a phenomenon where mutualistic relationship happened between plant and microbe in which the microbes live inside the plant without causing any symptoms or diseases (Wani et al. 2015). Endophytic bacteria was defined by Ryan et al. (2008) as a bacteria that occupy different areas in the plant tissue and does not cause pathogenicity. While Hallmann et al. (1997) stated that endophytic bacteria are bacteria that colonize latently or actively and locally or systematically of plant tissue. However, all of the definition is still very limited because it does not provide any information about the other types of symbiosis between endophytic bacteria and plants except beneficial relationship. Sturz et al. (2000) defined that endophytic bacteria are nonpathogenic bacteria that form various types of relationship with its host such as beneficial, neutral, or detrimental. From the previous definition, the endophytic bacteria can be summed as bacteria isolated from sterile surface tissue and colonize plant tissue both locally and systemically and form various types of with its host such as beneficial, neutral, or detrimental.
Based on Wani et al. (2015), endophyte can be divided into two categories: systemic and non-systemic endophytes (Table 1). Petrini (1991) has defined systemic endophyte as organisms that occupy the parts of plant tissue at least one part of their life cycle, perform a symbiotic relationship with its host, and does not cause any symptoms or certain diseases. While the non-systemic endophyte is endophyte that occupies the part of plant tissue in a relatively short time and form a different relationship with its host plant depend on environmental conditions (Salud et al. 2011).
Table 2 Endophyte classification based on the lifestyle (Hardoim et al. 2008)
Endophytic bacteria can come from various sources, but rhizosphere and phyllosphere are believed to be the main sources of endophytic bacteria (Hallm ann et al. 1997). It is indicated by the similarities of community existing in the plant tissue with community in the rhizosphere and phyllosphere. However, the further study stated that the rhizosphere community has the greater opportunities to occupy niches in the plant tissue compared by phyllosphere community. The small possibility to occupy a niche as endophyte in the plant tissue. It is because when the phyllosphere bacteria enter and colonize plant tissue, there will be competition between the bacteria that already exist in the plant tissue with the phyllosphere bacteria. Beside rhizosphere and phyllosphere, seeds and vegetative material are also believed to be the source of endophytic bacteria (Kaga et al.
2009).
Interaction between the candidates of endophytic bacteria with the plants had been initiated before the bacteria colonizing plant tissue. In conclusion, the success of endophytic bacteria to enter the plant tissue depend on some important
No Criteria Endophyte
Systemic Non-systemic 1 Relationship Form a mutualistic relationship
with the host
Form a various relationship with its host (mutualism, latent saprophyte, etc)
3 Colonization Systemic colonization in intercellular space
5 Coevolution Coevolution within the plant exist Coevolution within the plant rarely exist 6 Diversity Low diversity High diversity
Table 1 Endophyte classification based on colonization in the plant tissue (Wani et al. 2015)
No Criteria Endophyte
Facultative Obligate 1 Habitat Soil, surface of plant, inside the
plant tissue, or artificial nutrient
Inside the plant tissue
2 Host range Broad-host range Narrow-host range 3 Mode of
Transmission
Horizontal Vertical or use vector
4 Source of endophytic
Commonly from rhizosphere or phyllosphere
Exist in the plant tissue in a long period
5 Mode of Entrance
Commonly enter from wounds or natural openings
stages include pre-colonization, colonization, and post-colonization. Pre-colonization involves the finding and recognizing the host. Host finding is carried out by the movement of the candidate of endophytic bacteria towards plant tissue in various ways for example by chemotaxis. After finding a suitable host, candidate of endophytic bacteria will attach to the surface of host tissue and cause the host plasma membrane damage. After the attachment, the next stage is the introduction. Results from the introduction stage between candidates of endophytic bacteria and plants can be positive or negative. The introduction with a negative response (incompatible) between bacteria and the host will induce defense system in plants such as the induction of resistance, hypersensitivity, phytoalexin production, the destruction of the cell wall, formation of papillae, and so on. Meanwhile, if the introduction is positive (compatible), the plant will produce nutrients required for bacteria growth. The recognition process is a pre-selection of endophyte communities in the plant (Hallmann et al. 1997)
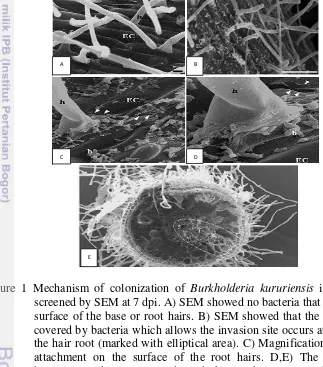
Colonization stage of endophytic bacteria in plant tissue (Figure 1) generally happened through natural opening such as lenticels and stomata, natural wound (due to biotic or abiotic factor), the emergence of lateral roots area,
A B
C D
E
A
Figure 1 Mechanism of colonization of Burkholderia kururiensis in rice plant screened by SEM at 7 dpi. A) SEM showed no bacteria that colonize the surface of the base or root hairs. B) SEM showed that the root surface covered by bacteria which allows the invasion site occurs at the base of the hair root (marked with elliptical area). C) Magnification of bacteria attachment on the surface of the root hairs. D,E) The invasion of bacteria into the root tissue through the root hairs. EC, epidermal cells; h, the root hairs ; b, bacteria (Mattos et al. 2008).
B
C D
epidermis conjugation, radicle germination, or also penetration using hydrolytic enzyme such as cellulase and pectinase (Hallmann et al. 1997). The penetration process of endophytic bacteria through natural openings happened either actively or passively. Penetration is done passively assisted by fluid flowing from the leaf through the stomata. While active penetration can be done by using hydrolysis enzyme. However, there are regulations that cause the enzyme is only produced when the penetration process, but after entering into the plant tissue, the enzyme is no longer produced. This is acceptable because if the enzyme is continuously produced, it will damage the plant tissue and create negative relationship between endophyte and its host. The patterns of endophytic bacteria colonization also vary depend on the strain and its species. Endophytic bacteria commonly colonize plant in the intercellular tissue. But others colonize the intracellular part, as well as vascular tissue. Interaction between endophytic bacteria and plants depends on the type of its endophytic bacteria.
Endophytic community structure is dynamic. It is influenced by several factors, both abiotic and biotic. Biotic factors include conditions in its host plants, such as the availability of nutrients, specificity, the stage of development, health, and etc. While the influences of abiotic consist of physical and chemical factors. Physical factors include temperature, rainfall, UV radiation, and moisture (Hallmann et al. 1997). While chemical factors include differences of the soil profile like pH, salinity, and other chemical compounds that may indirectly affect the composition of the bacterial communities in the rhizosphere which is considered as the main source of endophytic bacteria (Berg and Smalla 2009). Marschner et al. (2001) stated that the composition of the rhizosphere bacteria affected by complex interactions between soil type, plant species, and the location of the root zone. Ahlholm et al. (2007) also reported that the success of foliar endophytic bacteria infect the host plant depends on the interaction between the environment and the host genotype. It means that different environmental conditions can influence the selection of endophytic bacteria in plant tissue to survive in favorable environmental conditions.
Endophytic actinomycetes is a type of bacteria that recently developed mainly in the field of agriculture. Actinomycetes are a group of microorganisms that are most distributed in nature. In the natural habitat, Streptomyces is the most common actinomycetes group and often found on the total population of actinomycetes. Some genera of actinomycetes such as Actinoplanes, Amycolatopsis, Catenuloplanes, Dactylosporangium, Kineospora, Microbispora, Micromonospora, Nonomuraea are very difficult to be isolated and grown, usually referred to rare actinomycetes (Monisha et al. 2011). Some endophytic actinomycetes were reported to have antagonistic activity against various kinds of pathogens in rice (Tian et al. 2004; Hastuti et al. 2012). Streptomyces, Nocardia
Rice Plant
Rice (Oryza sativa L.) is an annual plant that has a fibrous roots and short stem which form leaf midrib to support the leaves. Rice is known as a source of carbohydrates, especially the endosperm. Other parts of rice commonly known as industrial raw materials for example the outer shell of rice (bran) as an oil, rice husk as fuel or material for paper and fertilizer. The taxonomical classification of the rice plant is as follows:
Kingdom : Plantae Patenggang, IR 64, Ciherang and Inpara 2 (Table 3). Selection of those varieties based on different types of agro-ecosystems. Situ Patenggang is commonly planted on dry land; IR 64 and Ciherang are cultivated on irrigated land; Inpara 2 is planted on tidal swampland.
Characteristics
Rice plant varieties
IR64 Ciherang Inpara 2 Situ Patenggang Age (days) ±115 116-125 ±128 ±124
Height (cm) ±85 107-115 ±103 ±134 Productive tiller (per
number of stem)
25 14-17 16 11
Shattering Tight Intermediate Intermediate Intermediate Resilience Tight Intermediate Intermediate Intermediate Amylose content 27 23 22,05 21,9
A wide variety of microorganisms, including fungi, actinomycetes, and other bacteria have been found in plants and referred to as endophyte. Endophytic bacteria that have been isolated from various parts of the rice plant for last 5 years by both culturable and unculturable methods can be seen in Table 4.
PCR-DGGE
DGGE (Denaturing Gradient Gel Electrophoresis) is a molecular technique used to separate DNA fragments from PCR products that have the same size of base pair but different sequence arrangement. It is separated by acrylamide gel with a gradient denaturant from low to high (Fischer and Lerman 1983; Myers
et al. 1987; Rosenbaum and Riesner 1987; Riesner et al. 1991). Separation of effectiveness to detect about 50% of the sequences in the DNA fragment until 600 bp, but using GC-clamp, the percentage of effectiveness can be increased up to 100% (Myers et al. 1985; Sheffield et al. 1989). GC-rich sequences will prevent an open double-stranded DNA totally becomes single stranded (Sheffield et al. 1989; Sheffield et al. 1992).
Table 4 Some endophytic bacteria isolated from rice plant
Rice Part Bacteria Taxon
(Bacteria and Actinomycetes) Rice Species Ref. Grains, root,
Acinetobacter oryzae sp. nov. Oryza alta Chaudhary et al 2011
Seed Burkholderia cepacia, Citrobacter
sp., Citrobacter sp., Citrobacter sp., Bacillus amyloliquefaciens, B. amyloliquefaciens,
and B. thuringiensis
Oryza sativa L. Gangwar et al. 2011
Root Burkholderia cepacia Oryza sativa L. Singh et al. 2011 Root Streptomyces sp. GMKU 3100 Oryza sativa L.
cv.KDML105 Rungin et al. 2012 Leaves Pantoea ananatis and Pseudomonas syringae Oryza sativa L. Ferrando et al. 2012
Root Stenotrophomonas maltophilia Oryza sativa L. Zhu et al. 2012 Root Pesudomonas sp.,Bacillus, Azotobacter,
and Enterobacter
Oryza sativa L. Narayanasamy 2012
Leaves, stems, and roots
Penibacillus, Microbacterium, Bacillus, and
Klebsiella.
Oryza sativa L. Ji et al. 2014
Seed Staphylococcus, Rhizobium,
Microbacterium and Methylobacterium.
Oryza sativa L. Jiang et al. 2013
Seed Pantoea sp. Sd-1 Oryza sativa L. Xiong et al. 2014 Root Rhizobium rhizoryzae sp. nov. Oryza sativa L. Zhang et al. 2014
Effective staining for DGGE gel is by using SYBR green (Muyzer et al. 1997). Benefits of using SYBR green is reducing background staining on the gel, so it can facilitate band profile both dominant and less dominant.
The principle of DGGE is the using of gradient to separate DNA fragments. Good separation of DNA fragments can be done by optimizing the gradient and electrophoresis time. Sequence variations would lead to the differences in melting point and the position of the stop migration in the gel. When the lowest melting point is reached then a portion of the double-stranded DNA will be opened and the migration process will immediately stop. Melting behavior of DNA fragments can be observed by using perpendicular gradient gel. Gel perpendicular has a denaturant gradient increase from left to right, perpendicular towards the electrophoresis direction (Fischer and Lerman 1994). The electrophoresis proceeded for 16 hours at 100V. The optimal time of electrophoresis is determined by the electrophoresis gradient. While the parallel gradient gel is the gel that has an increased gradient from top to bottom, parallel to the electrophoresis direction. DGGE is also useful for proceeding many samples at the same time. Apparatus of DGGE can be obtained by different commercial companies such as Bio-Rad (Herculas, USA).
DGGE technique has several advantages such as simple, easy to use for regular laboratory, and the results are also easy to interpret. In its application to the study of ecology, DGGE can be used to study the changes of the community because it can analyze many different samples simultaneously based on environmental change. In addition, this method can be used to observe the richness of bacterial isolation, for example to analyze the results of the PCR product of pure cultures whether the product contains one or more fragments. Another benefit is to compare the different extraction protocol by comparing the ability of producing different 16S rRNA fragment from different DNA extraction protocol (Heuer and Smalla 1997; Liesack et al. 1997). It is also can be used to screen the clone library in a suitable vector (Kowalchuk et al. 1997). Determining PCR and colony bias to know the error rate of the DNA polymerase in DNA synthesis (Keohavong and Thilly 1989).
DGGE has a limitation in community screening because it can proceed the sample with maximum size around 600 bp, so it does not contain a lot of information for accurate identification. Application of GC clamp in the PCR process is also sometimes producing primer dimers so variable of coloring gel can be reduced.
METHODS
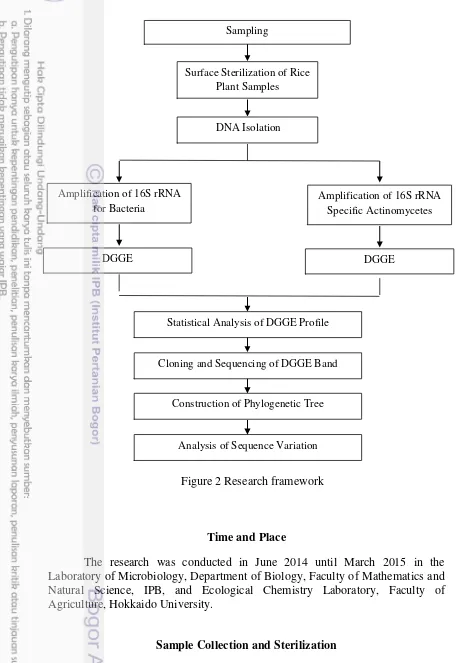
Research Framework
Time and Place
The research was conducted in June 2014 until March 2015 in the Laboratory of Microbiology, Department of Biology, Faculty of Mathematics and Natural Science, IPB, and Ecological Chemistry Laboratory, Faculty of Agriculture, Hokkaido University.
Sample Collection and Sterilization
Sampling site was conducted in Bogor and Cianjur, West Java, Indonesia. The part of plant samples (root, stem, and leaf) were collected from four healthy
Figure 2 Research framework Surface Sterilization of Rice
Plant Samples
DNA Isolation
Amplification of 16S rRNA for Bacteria
Amplification of 16S rRNA Specific Actinomycetes
DGGE DGGE
Cloning and Sequencing of DGGE Band
Construction of Phylogenetic Tree Sampling
Statistical Analysis of DGGE Profile
rice cultivars (IR 64, Inpara 2, Situ Patenggang, and Ciherang) at vegetative state (30 days old) (Mahyarudin 2014). The characteristic of agricultural soil of the sampling site has been analyzed from previous study (Mahyarudin 2014) and presented in Table 5 and 6. Surface sterilization of the samples was conducted based on Coombs and Franco (2003), with modification. The samples were washed by tap water to clean the surface part. About 0.5-1 g of each samples were immersed in 70% ethanol solution for 1 minute, washed with sodium hypochlorite (NaOCl) 1% for 5 minutes, rinsed with 70% ethanol for 1 minute, and finally washed three times with sterile distilled water. Surface sterilization was validated by spreading the last washing water on culture media and incubated for 1 month. The samples that did not show any contamination were used for further analysis.
Table 6 Characteristics (II) of agricultural soil from sampling site (Mahyarudin 2014)
DNA Isolation
Table 7 List of the primers used in this study
No System Primer name Sequence (5'->3') Ref. 1 Select for
actinomycetes
16Sact1114R GAGTTGACCCCGGCRGT Martina et al. 2008
2 Universal 27F AGAGTTTGATCCTGGCTCAG Martina et al. 2008
3 Universal P338F ACTCCTACGGGAGGCAGCAG Overeas et al. 1997
4 Universal P518R ATTACCGCGGCTGCTGG Overeas et al. 1997 5 DGGE P338F-gc CGCCCGCCGCGCGCGGCGGG
CGGG
GCGGGGGCACGGGGGGACTC CTAC
GGGAGGCAGCAG
Overeas et al. 1997
6 Select for Bacteria 799F-mod3 CMGGATTAGATACCCKGG Hanshew et al. 2013
7 Universal 1389R ACGGGCGGTGTGTACAAG Hongoh et al. 2003
isopropanol 1.5x volume of supernatant. After that, the mixture was homogenized for 30 seconds. The total of 700 mL of the mixture was transferred to the GD column that has been put on the collection tube, then centrifuged at 13000 rpm for 2 minutes. Supernatant in the collection tube was discarded. The process was repeated until the mixture in microtube dried. The total of 400 mL W1 buffer was added to the GD column and centrifuged at 13000 rpm for 30 seconds. Supernatant in the collection tube was discarded. After that, 600 mL washing buffer was added to the GD column and centrifuged at 13000 rpm for 30 seconds. GD column was centrifuged again at 13000 rpm for 3 minutes to dry the matrix volume. GD column that has been dried was put on the 1.5 mL microtube. A total of 30 mL of elution buffer was added to the GD column and allowed up to 20 minutes and then centrifuged for 30 seconds at 13000 rpm for obtain total DNA. The results of DNA isolation was electrophoresed on 1% gel agarose and visualized.
Amplification of 16S rRNA Gene-specific Bacteria and Actinomycetes
PCR amplification of bacteria domain from root, stem, and leaf was done using 799F-mod3/1389R primer set to amplify fragment size ±600 bp. The reaction mixture was as follows: 10 µL Ampli-Taq Gold 360 (Applied Biosystems, Carlsbad, CA), 0.4 µL of each primer (10 µmol), 1 µL of DNA template (30 ng), and 8.2 µL sterile MiliQ water. The amplification was performed in TAKARA Thermal Cycler (Takara, Dalian, China) using initial denaturing of 1 minute at 95 oC, followed by 30 cycles of 30 seconds denaturing at 95 oC, 1 minute annealing at 55 oC, and 1 minute extension at 72 oC, completed by 7 minutes final extension at 72 oC.
DNA template (30 ng), and 8.2 µL sterile MiliQ water. The amplification was carried out using initial denaturing of 5 minutes at 94 oC, followed by 30 cycles of 1 minute denaturing at 94 oC, 30 seconds annealing at 55 oC, and 30 seconds extension at 72 oC, completed by 3 minutes final extension at 72 oC.
Analysis and Cloning of DGGE Bands
DGGE was performed using Bio-Rad DCode system (Bio-Rad, Hercules, CA, USA) in 6% and 8% (w/v) polyacrylamide gel (acrylamide-bisacrylamide, 37.5:1) with 30% to 70% denaturing concentrations for bacteria and actinomycetes DGGE analysis, respectively (100% denaturant corresponding to 7M urea and 40% deionized formamide). Electrophoresis was performed at 100 V and 60 °C for 16 hours in 1 × Tris-acetate-EDTA (TAE). The gel was stained by 15 µL of Sybr Gold dye (Molecular Probes, Invitrogen, Cergy Pontoise, France) in 1 × TAE buffer (150 mL) for 30 minutes with dark conditions. Gel was screened by using Typhoon imaging system (Amersham, Piscataway, NJ, USA). Band profile image was analyzed using Phoretix 1D software (Nonlinear Dynamics, Newcastle, UK) to estimate the total bands that appeared on polyacrylamide gel.
Statistical analysis of DGGE profile was conducted by using alpha diversity (Shannon-wiener/He) to estimate the diversity within each sample and beta diversity (Dice similarity coefficient/SD) to estimate the similarity of band pattern between the samples. The quantification of statistical analysis was conducted using PAST Software(Hammer et al. 2001) based on the estimation analysis of band intensity using Phoretix 1D software (Nonlinear Dynamics, Newcastle, UK). The index was calculated by following equation:
Where, Na represented the number of bands detected in sample a; Nb represented the number of bands detected in sample b; Nc represented the number of bands detected in both samples; s represented the number of species in the sample; Pi represented the proportion of species i in the sample (Han et al. 2014).
Cloning steps was done to purify the DGGE fragments. First phase was
preparation of competent cell (E.coli). The total of 100 mL of LB medium was prepared by adding 2.5 g LB in 100 mL MiliQ water, then sterilized at a temperature of 121 oC for 20 minutes. After that, the cells were inoculated into LB media and grown by shaking the culture overnight with 200 rpm at 37 °C. After that, 2 mL of the culture was inoculated into 20 mL LB and shook for 2-3 hours at 37 °C with 200 rpm. The total of 1 mL culture was harvested and centrifuged at 8000 rpm for 1 minute at 4 °C. The supernatant was discarded and 1 mL of cold 0.1 M CaCl2 was added to the pellet and re-suspended slowly. After that, the
CaCl2 was added to the pellet and re-suspended slowly, then incubated on ice for
20 minutes. After incubation, the culture was centrifuged at 8000 rpm for 1 minute at 4 °C. The supernatant was discarded and about 100 mL of CaCl2 was added to the pellet and stored until used.
Second phase was ligation. Reaction in the ligation step was done by mixing 5 µL rapid 2x ligation buffer, T4 DNA Ligase, 1 µL pGEM-T vector (Promega, Madison, WI, USA) (50ng), 3 µL of fresh PCR product, 1 µL of T4 DNA ligase (Promega, Madison, WI, USA) and nuclease free water until it reached the final volume 10 µL.The reaction was mixed by pipetting techniques. The reaction was incubated overnight at 4 °C.
Third phase was transformation. The total of 2 µL Ligation mix was added to the 50 µL competent cells. After that, the mixture was incubated on ice for 15-20 minutes. Then the mixture was put in the heat shock for 90 seconds at a temperature of 42 oC. Cells were inoculated on LB solid media that has been added by ampicillin and X-Gal and incubated overnight at 37 °C. White colonies grown on the medium were transferred to the new solid LB media to be used as PCR template stock colony.
Forth phase was PCR colony. The total of 1 ose white colonies was inoculated in a PCR reaction with 30 µL volume of 15 µL Ampli-Taq Gold 360 (Applied Biosystems, Carlsbad, CA), 0.6 µL (10 µmol) of each primer, and 13.8 µL of sterile MiliQ water. PCR conditions used consisted of pre-denaturation step (95 °C, 5 minutes), denaturation (95 °C, 30 seconds), annealing (55 °C, 45 seconds), extension (72 °C, 1 minute), and a final extension (72 °C, 5 minutes). PCR process is done for about 30 cycles. Visualization of PCR products was done by electrophoresis in agarose gel 1% (w/v).
Sequencing of the 16S rRNA Gene and Construction of Phylogenetic Tree
Sequencing of 16S rRNA gene performed using ABI 3730 automated sequencer (Invitrogen, Shanghai, China). PCR products about 20 µL was purified by using a simple column. A total of 100 µL of TE buffer + G25 were added to the column and centrifuged. The supernatant from the centrifugation was disposed from microtube. Total of 20 µL PCR product was added to the column and centrifuged. The resulting of supernatant that has been centrifuged was stored for PCR sequencing.
Sequencing PCR reactions containing 3.8 µL of 5 x buffer, 3.2 µL of each primer (1 µmol), 2 µL premix (4x dilution), 1 µL of DNA template, and 10 µL of sterile MiliQ water. PCR condition used was 96 oC for 5 minuts, 30 cycles for the condition of 96 oC for 30 seconds, 50 °C for 15 seconds, and 60 °C for 4 minutes.
the sequencer machine. Results were compared to the GenBank nucleotide
sequence database of NCBI BLAST.N through the website
(http://www.ncbi.nlm.nih.gov/BLAST).
The sequencing results were compared to the GenBank nucleotide
sequence database of NCBI BLAST.N through the website
(http://www.ncbi.nlm.nih.gov/BLAST). Relationship analysis among the sequenced DNA was performed based on phylogenetic tree using the neighbor-joining method and software MEGA 5.0 and evaluated using bootstrap analysis with 1000 replication (Tamura 2011)
Sequence Variation Analysis
Sequence variation analysis was conducted to analyze the differences of nucleotides between different groups of bacteria and actinomycetes. Sequence analysis was performed to DGGE bands that have been determined its species affiliation. Sequences were aligned using MEGA 5.0 software (Tamura 2011) and the variable region proceeded further to analyze the patterns differences among the groups.
Specifications of species (DGGE band) originated from Indonesian rice plants were analyzed by comparing the bands that have same affiliation with its species affiliation. Specific nucleotide sequences that exist only on DGGE bands were selected and analyzed further.
RESULT AND DISCUSSION
Result
Effectiveness of Surface Sterilization
Confirmation of surface sterilization effectiveness was done using HV and NA media. After 30 days of incubation, no bacteria or actinomycetes showed in the media. It indicated that the effectiveness of surface sterilization was good.
DNA Isolation
Table 8 The quantity and quality of DNA extracted from rice plant tissue
Note: (R) Root, (S) Stem, (L) Leaf
Endophytic Bacterial Community in Indonesian Rice Cultivars
Bacterial communities in root, stem, and leaf of rice plant were analyzed using primer set of 799F-mod3/1389R which specifically amplified bacteria domain with percent of chloroplast read about 0.88 (Hanshew et al. 2013) resulted the fragment size around ±600 bp (Figure 3).
The structures of endophytic bacterial communities of four Indonesian rice cultivars (root, stem, and leaf) were compared by DGGE analysis. A total of 17 bands were excised from DGGE gel (Figure 4a). The present of the bands varied among the samples in which no common band detected in all samples. Some of the bands were only found in specific sample i.e. band 7, 19, and 20 were only found in IR64 rice cultivar, while band 21 was only found in Inpara 2 rice cultivar. There was different number of bands among the samples in which
No Sample
Nucleic Acid concentration
( ng/µL)
A260 A280 260/280 260/230
1 R.Ciherang 32.4 0.648 0.38 1.7 0.47 2 S Ciherang 26.2 0.525 0.27 1.95 1.12 3 L Ciherang 11.5 0.231 0.115 2.01 1.07
4 R.IR64 39.8 0.795 0.688 1.16 0.12
5 S.IR64 15.8 0.315 0.255 1.23 0.13
6 L.IR64 31.6 0.631 0.373 1.69 0.44
7 R.Inpara2 28.3 0.566 0.298 1.9 1.14 8 S.Inpara2 12.6 0.251 0.137 1.83 1.43 9 L.Inpara2 14.2 0.284 0.164 1.73 1
10 R.SP 13,0 0.26 0.146 1.78 1.01
11 S.SP 12.2 0.244 0.129 1.89 1.04
12 L.SP 10.4 0.209 0.097 2.15 1.49
Figure 3 PCR amplification of 16S rRNA gene specific bacteria from four Indonesian rice plant cultivars with product size ±600 bp using 799F-mod3/1389R. R) Root; S) Stem; L) Leaf
17
a b
2.199 Indonesian rice cultivars for endophytic bacterial community using primer set 799F-mod3/1389R
Ciherang and IR64 showed high abundance of bands, while Inpara 2 and Situ Patenggang showed low abundance of bands.
Shannon-wiener (He) analysis were used in this study to estimate the microbial diversity in each sample, in which high index value referred to the high diversity of species (bands) within a sample. The result for bacterial community (Figure 5) showed He index value of IR64 and Ciherang (1.855-3.011) were higher compared with Inpara 2 and Situ Patenggang (0.501-2.063). Unlike alpha-diversity (Shannon-wienner), the Dice similarity coefficient (SD) analysis was used to explain the similarity of species (band) composition between different types of samples. SD index value approach to 1 indicated high similarity of the structure composition between the samples compared. The result for endophytic
Table 9 Dice similarity coefficient analysis of endophytic bacterial community
bacteria (Table 9) showed the community structure in IR64 has a high similarity with Ciherang with SD index value 0.588-0.974. In addition, SD index value of Inpara 2 and Situ Patenggang shared low similarity of community structure if they compared with another samples with SD index value 0.167-0.615.Cluster analysis was done to estimate the order of relationship among the samples. Cluster analysis of bacterial community (Figure 6) showed that IR64 and Ciherang have a close cluster, while Inpara 2 and Situ Patenggang grouped into the other cluster.
Phylogenetic analysis for bacterial community was done by selected 17 representative DGGE bands to be sequenced (Table 10). The result (Figure 7) showed the sequences of bands were grouped into 5 classes, which consisted of Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, Bacilli, and Flavobacteria. Bands 1, 2, 5, 6, 10, 11, 12, and 13 were grouped into Gammaproteobacteria affiliated to genera Cellvibrio, Pseudomonas and
Escherichia. Band 7 was grouped into Betaproteobacteria affiliated to genus
Burkholderia. Band 9 was grouped into Alphaproteobacteria which affiliated to genus Brevundimonas. On the other hand, bands 3 and 15 were grouped into class Bacilli affiliated to genus Sporosarcina. Bands 14, 16, and 17 were grouped into class Flavobacteria affiliated to genera Myroides and Flavobacterium. Meanwhile, the sequence of band 8 was identified as an archaeon. Bands 5, 15, and 16 found only in IR64 rice cultivar showed the highest homology with Pseudomonas putida
strain ZJUTBX04, Brevibacillus brevis strain DZBY12 , and Myroides odoratus
strain S5 respectively. Band 17 detected only in Inpara 2 showed the highest homology with Flavobacterium ceti strain 454-2.
Table 10 Phylogenetic affiliation of endophytic bacterial community
Band Species affiliation Similarity Base Position
Accession
number Group 1 Cellvibrio japonicus strain
UEDA 107
97% 819-1335 NR_074804.1 Gammaproteobacteria
2 Cellvibrio japonicus strain UEDA 107
5 Pseudomonas putida strain ZJUTBX04
98% 812-1331 JF682514.1 Gammaproteobacteria
6 Escherichia fergusonii
strain NBRC
98% 857-1347 NR_114079.1 Gammaproteobacteria
7 Burkholderia cepacia strain LO6
93% 912-1316 CP011301.1 β-proteobacteria
8 Thaumarchaeota archaeon
SAT1
98% 751-1286 CP011097.2 Archae
9 Brevundimonas olei Strain DUCC378
92% 728-1224 KP318465.1 α-proteobacteria
10 Pseudomonas poae
RE*1-1-11 Cellvibrio mixtus strain J3-8 96% 778-1337 KC329916.1 Gammaproteobacteria
Cellvibrio mixtus strain ACM 2601
96% 779-1338 NR_041884.1 Gammaproteobacteria
12 Cellvibrio mixtus strain ACM 2601
90% 779-1338 NR_041884.1 Gammaproteobacteria
Cellvibrio mixtus strain J3-8 90% 778-1337 KC329916.1 Gammaproteobacteria 13 Pseudomonas
brassicacearum subsp.
brassicacearum NFM421 strain NFM421
99% 777-1356 NR_074834.1 Gammaproteobacteria
14 Myroides odoratus strain S5 99% 695-1254 KM268942.1 Flavobacteria
Myroides odoratus strain NBRC 14945
99% 746-1305 NR_112976.1
15 Brevibacillus brevis strain DZBY12
97% 796-1333 KM191288.1 Bacilli
16 Myroides odoratus strain S5 91% 695-1254 NR_112976.1 Flavobacteria
Myroides odoratus strain NBRC 14945
91% 746-1305 KM268942.1 Flavobacteria
17 Flavobacterium ceti strain 454-2
Analysis of variation pattern has been done on the sequences of DGGE bands with total size 437 bp. Position of sequences ranges in ± 315 -547 (Figure 8) in the region of 16S rRNA gene (± 1500 bp) within the conservation area V4, V5, V6, V7, and V8. Bands were grouped into several classes and genera based on the results of NCBI blast. The result showed 103 base position has a specific pattern in bacterial group (Appendix 3). In class level, specific pattern consisted of 48 specific base sites of Gammaproteobacteria, 68 specific base sites of Bacilli, and 67 specific base sites of Flavobacteria. While, in genus level, specific pattern consisted of 83 specific base sites of Cellvibrio, 94 specific base sites of
Pseudomonas, 98 specific base sites of Sporosarcina, and 99 specific base sites of
Myroides.
The bands that have the same affiliation to one species were compared to analyze whether there are specific pattern of bacteria originated from Indonesian rice plant with the affiliate bacteria isolated from different sample (Table 11). The results showed the presence of several base variations (single nucleotide polymorphism) specifically exist in the bands from Indonesian rice plant. Some of variation showed in Sporosarcina contaminans strain CCUG 53 915,
Sporosarcina koreensis strain F73, Pseudomonas brassicacearum subsp. Figure 7 Phylogenetic Tree of bacterial 16S rRNA gene-targeted DGGE band
brassicacearumstrain AF129, Cellvibrio japonicus strain Ueda107, and Cellvibrio mixtrus strain ACM 2601. Comparison of the base variations can be seen in the table . Clone band 1 and 2 affiliated with Cellvibrio japonicus strain Ueda107 from outside of Indonesia has 10 specific base sites. Clone band 11 and 12 have 1 specific base site compared with Cellvibrio mixtrus strain ACM 2601. No specific base site in clone 14 and 15 compared with Myroides odoratus strain S5 and
Myroides odoratus strain NBRC 14945. There were 4 specific base sites among clone 3 and 4 compared with Sporosarcina contaminans strain CCUG 53 915 and
Sporosarcina koreensis strain F73. No specific site found in clone band 10 and 13 compared with Pseudomonas brassicacearum subsp. brassicacearumstrain
AF129.
437 bp
Figure 8 Position of DGGE band region amplified by 799F-mod3/1389R primer set within complete sequence of 16S rRNA in the GenBank data sequence
17
16S rRNA Region 1
2 3
4
5
6 7 8
9 10
11 12
13 14 15
Endophytic Actinomycetes Community in Indonesian Rice Cultivars
PCR amplification for actinomycetes community was done by 2 stages PCR. The first stage used specific primer for actinomycetes 27F/16Sact1114R resulted fragment size ±1080 bp (Figure 9a). First PCR product was used as a template for second PCR resulted fragment size ±195 bp (Figure 9b).
a b
Figure 9
The structures of special bacterial taxon, endophytic actinomycetes, of four Indonesian rice cultivars (stem and leaf) were also compared by DGGE analysis. Total of 16 bands were excised from DGGE gel (Figure 10). Band profiles of actinomycetes community showed some common bands appearing in all of the samples i.e. band 3, 4, 8, 10, and 16. Band 12 was only found in Inpara 2 rice cultivar. The DGGE profile showed not highly different number of bands among the samples.
The result for actinomycetes community (Figure 11) showed He index value was not too different (2.272-2.925) suggested the diversity structure among the samples were almost similar. Meanwhile, the result for endophytic actinomycetes (Table 12) showed the community structure among the samples shared relatively high similarity with SD index value 0.563-0.778, except for comparison of community structure between Ciherang and Situ Patenggang that shared low similarity with SD index value 0.387-0.48. Cluster analysis was done to estimate the order of relationship among the samples. Cluster analysis of actinomycetes (Figure 12) showed that IR64 and Ciherang have a close cluster, while Inpara 2 and Situ Patenggang grouped into the other cluster
Figure 9 PCR amplification of 16S rRNA gene specific actinomycetes from four Indonesian rice plant cultivars. a) First step PCR with product size ±1080 bp using 27F/16Sact1114R. b) Second step PCR with product size±195 bp using 338F/518R
25
a b
Figure 100 Actinomycetes community in four Indonesian rice cultivars. a) PCR-DGGE fingerprinting b) Illustration of DGGE band using 1D Phoretix software showed 1-16 excised bands. S (Stem); L (Leaf)
.
Figure 12 Hierarchical cluster analysis result of actinomycetes DGGE profiles demonstrated graphically as an UPGMA dendrogram (p-distance)
Figure 11 Shannon diversity index of 16S rRNA DGGE profiles of four Indonesian rice cultivars for endophytic actinomycetes community using second PCR primer set 338F-gc/518R
Table 12 Dice similarity coefficient analysis of endophytic actinomycetes community
Samples 1 2 3 4 5 6 7 8
S-Ciherang
L-Ciherang 0.706
S-IR64 0.722 0.588
L-IR64 0.778 0.706 0.778
S-Inpara 2 0.571 0.606 0.743 0.743
L-Inpara 2 0.579 0.611 0.579 0.737 0.649
S-Situ P 0.467 0.429 0.600 0.600 0.690 0.563
L-Situ P 0.485 0.387 0.667 0.667 0.750 0.686 0.741
Table 13 Phylogenetic Affiliation of Endophytic Actinomycetes Community
Band Species affiliation Maximum identity
Base Position
Accession
number Group 1 Cellulomonas flavigena strain
DSM 20109
100% 373-504 NR_074490.1 Cellulomonadaceae
2 Kocuria polaris strain CMS 76or 100% 373-506 NR_028924.1 Micrococcaceae
Kocuria rosea strain DSM 20447 100% 365-498 NR_044871.1 Micrococcaceae 3 Kocuria rosea strain DSM 20447 100% 365-498 NR_044871.1 Micrococcaceae
Kocuria polaris strain CMS 76or 100% 373-506 NR_028924.1 Micrococcaceae 4 Kocuria aegyptia strain YIM
70003
99% 374-512 NR_043511.1 Micrococcaceae
Arthrobacter aurescence TCI strain TCI
99% 357-487 NR_074272.1 Micrococcaceae
Arthrobacter ramosus strain CCM 1646
99% 360-497 NR_114963.1 Micrococcaceae
Arthrobacter arilaitensis strain Re117
99% 374-505 NR_074608.1 Micrococcaceae
Citricoccus nitrophenolicus strain PNP1
99% 370-502 NR_117546.1 Micrococcaceae
5 Microbacterium luteolum strain IFO 15074
obacterium luteolum
99% 333-471 NR_024636.1 Microbacteriaceae
6 Kocuria aegyptia strain YIM 70003
100% 374-512 NR_043511.1 Micrococcaceae
Arthrobacter aurescence TCI strain TCI
100% 357-487 NR_074272.1 Micrococcaceae
Arthrobacter ramosus strain CCM 1646
100% 360-497 NR_114963.1 Micrococcaceae
Arthrobacter arilaitensis strain Re117
100% 374-505 NR_074608.1 Micrococcaceae
Citricoccus nitrophenolicus strain PNP1
100% 370-502 NR_117546.1 Micrococcaceae
7 Microbacterium insulae strain DS-66
99% NR_044440.1 Microbacteriaceae
8 Kocuria aegyptia strain YIM 70003
99% 374-512 NR_043511.1 Micrococcaceae
Arthrobacter aurescence TCI strain TCI
99% 357-487 NR_074272.1 Micrococcaceae
Arthrobacter ramosus strain CCM 1646
99% 360-497 NR_114963.1 Micrococcaceae
Arthrobacter arilaitensis strain Re117
99% 374-505 NR_074608.1 Micrococcaceae
Citricoccus nitrophenolicus strain PNP1
99% 370-502 NR_117546.1 Micrococcaceae
9 Micrococcus luteus strain NCTC 2665
99% 349-487 NR_075062.2 Micrococcaceae
10 Calothrix desertica strain PCC 7102
85% 433-474 NR_114995.1 -
Thermodesulfovibrio yellowstonii
strain DSM 11347
79% NR_044075.1 -
Acinetobacter baylyi strain B2 98% NR_115042.2 -
11 Streptomyces acidiscabies strain ATCC 49003
99% 355-487 NR_116534.1 Streptomycetaceae
Streptomyces chartreusis strain ISP 5085
99% 315-448 NR_114825.1 Streptomycetaceae
Streptomyces scopiformis strain NBRC 100244
99% 342-547 NR_112586.1 Streptomycetaceae
12 Kocuria aegyptia strain YIM 70003
100% 374-512 NR_043511.1 Micrococcaceae
Arthrobacter aurescence TCI strain TCI
100% 357-487 NR_074272.1 Micrococcaceae
Arthrobacter ramosus strain CCM 1646
Band Species affiliation Maximum identity
Base Position
Accession
number Group
Arthrobacter arilaitensis strain Re117
100% 374-505 NR_074608.1 Micrococcaceae
Citricoccus nitrophenolicus strain PNP1
100% 370-502 NR_117546.1 Micrococcaceae
13 Citricoccus nitrophenolicus strain PNP1
100% 370-502 NR_117546.1 Micrococcaceae
14 Streptomyces chartreusis strain ISP 5085
98% 355-487 NR_114825.1 Streptomycetaceae
Streptomyces acidiscabies strain ATCC 49003
98% 315-448 NR_116534.1 Streptomycetaceae
Streptomyces scopiformis strain NBRC 100244
98% 342-547 NR_112586.1 Streptomycetaceae
15 Streptomyces chartreusis strain ISP 5085
98% 355-487 NR_112586.1 Streptomycetaceae
Streptomyces acidiscabies strain ATCC 49003
98% 315-448 NR_114825.1 Streptomycetaceae
Streptomyces scopiformis strain NBRC 100244
98% 342-547 NR_112586.1 Streptomycetaceae
16 Kocuria polaris strain CMS 76or 100% 373-506 NR_028924.1 Micrococcaceae
Kocuria rosea strain DSM 20447 100% 365-498 NR_044871.1 Micrococcaceae
Phylogenetic analysis for actinomycetes community (Table 13) showed cluster with 4 large families (Figure 13), which consisted of Microbacteriaceae, Streptomycetaceae, Cellulomonadaceae, and Micrococcaceae. Bands 5 and 7