PHYLOGEOGRAPHY OF TROPICAL EELS
(
Anguilla spp
) IN INDONESIAN WATERS
MELTA RINI FAHMI
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
STATEMENT OF DISSERTATION
AND INFORMATION SOURCE
Hereby I express that the dissertation entitled: “
Phylogeography of
Tropical Eel (
Anguilla spp
) in Indonesian Water
” is the original result of
my research and has never been submitted to obtain a Doctorate in similar
mayor at other universities. All of data and information that I had provided
in this dissertation are based on evidence and available references.
Bogor, March 2013
SUMMARY
MELTA RINI FAHMI. Phylogeography of Tropical Eel (Anguilla spp) in Indonesia Waters. Supervised by DEDY DURYADI SOLOHIN, PATRCIK BERREBI, KADARWAN SOEWARDI and LAURENT POUYAUD
The freshwater eels (Anguilla spp: Anguillidae) are popular as a commercial important food, because of good nutritional value. These fish are also a well known for their unique catadromous life histories. These species breed far from offshore after migrate thousands kilometers from their growth habitats in freshwater and estuarine to their spawning area in oceanic waters. Most of the investigations concerning eels are concentrated on temperate species, in the north hemisphere mainly because of the economic importance of these species. Nowadays, the population of temperate eel, dramatically decrease is caused by habitat damage, illegal fishing and climatic changes in the ocean. As a consequence, tropical eels become important eel nowadays in the market, as well as the research on tropical eels become a new challenge. One of primary problems in tropical eels is that they have overlapping range of most morphological character, so species identification on this genus are no more sufficient. Then molecular approaches have been proposed for eel identification.
This study has four main objectives: (1) to establish quick methods of identification of tropical eels; (2) to understand exact distribution and dispersal of tropical eel in Indonesia water; (3) to reveal the phylogenetic relationship among population in Indonesia and (4) to uncover population genetic structure of Anguilla bicolor between two Ocean. All of information obtained in this study revealed the conservation management of tropical eel in Indonesia water.
The semi-multiplex method was proposed in this study has demonstrated the efficiency for in identifying seven species and sub-species of tropical eels with only one step PCR. By using this method, one could reduce the number of necessary sequences while the results are very sure for each species determination (we easily identified 1112 specimens). All of species were obtained in this study showed overlapping morphology and distribution. Especially for small specimens (mainly glass eels), the molecular method appears as indispensable. This method has proven to be most simple, quicker, lower cost (no acrilamide migration), specific/sensitive, and highly reliable way.
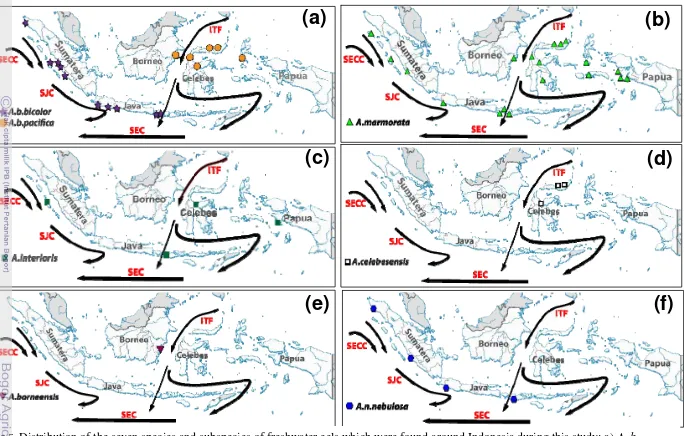
The geographic distribution of tropical eels of the genus Anguilla in Indonesian waters was establish by identified genetically of all sample that obtain during this study. The genetically identification applied the semi-multiplex PCR method that recently developed in this study. We recognized four species and subspecies with wide distribution: A. b. bicolor, A. b. pacifica, A. marmorata and A. interioris, two species with limited distribution, close to endemism: A. celebesensis and A. borneensis and one subspecies A. nebulosa nebulosa that is only spread in river flowing into Indian Ocean.
The economic important shortfinned eel, Anguilla bicolor, has a relatively wide geographic distribution compared to the 19 species and subspecies of genus Anguilla, it is distributed longitudinally from the eastern coasts of Africa through the seas around Indonesia to New Guinea in Pacific Ocean. The genotypes of seven microsatellite DNA were analysed for 180 specimens collected from 10 representative location where two subspecies have been found. Analysis with seven microsetallite loci showed Expected (He) and observed (Ho) heterozigosities of each locus range from 0,594 to 0,921 and from 0,250 to 1,000 respectively. All off locus showed high polymorphic. Based on FST value and
clustering test, there is no structure and fragmentation of A. bicolor in Indonesia water.
A critical first step in species conservation is to gain a clear understanding of its taxonomy. Considering Indonesian waters that are inhabited by several sympatric species of tropical eels, almost all morphological character in this genus are overlapping. So a rapid and efficient identification technique is needed. Semi multiplex PCR that has been developed in the present study has successfully distinguished seven species that inhabit the waters of Indonesia through one-step PCR. After established the identification species subspecies methods then we constructed distribution and dispersal pattern, phylogenetic relationship among species that inhabit Indonesian water and population genetic structure of species that have widespread distribution. All of information obtained in this study was needed for conservation management of anguillid in Indonesian water.
Key words: Tropical eel, Anguilla spp, semi-multiplex PCR, Indonesia water and
Copyright©2013 by Bogor Agricultural University Copyright are protected by law,
1. It is prohibited to cite all or part of this thesis/ dissertation without referring to and mentioning the source.
a. Citation only permitted for sake of education, research, scientific problem.
b. Citation doesn’t inflict the name and honor of Bogor Agricultural University.
Dissertation’s title : Phylogeogphy of Tropical Eels (Anguilla spp) In
Indonesian Waters
Name : Melta Rini Fahmi
Student ID : G 362080061
Departement : Biology
Aproved by the Supervisor Commite
Dr. Dedy Duryadi Solihin Head
Prof. Dr. Patrick Berrebi Prof. Dr.Kadarwan Soewardi, M.Sc
Member Member
Dr.Laurent Pouyoud M.Sc Member
Acknowledged by:
Head of Biology Programme of Dean of Graduate School Graduate School Bogor Agricultural University
TABLE OF CONTENTS
Page
TABLE OF CONTENTS ... i
LIST OF TABLES ... iii
LIST OF FIGURES ... iv
LIST OF APPENDIX ... v
I. GENERAL INTRODUCTION Background ... 1
Questions to Solve ... 7
Framework ... 7
II. A NOVEL SEMI-MULTIPLEX PCR ASSAY FOR IDENTIFICATION OF TROPICAL EEL GENUS Anguilla IN INDONESIAN WATER Abstract ... 10
Introduction ... 10
Material and Methods ... 12
Result ... 16
Discussion ... 18
III. DISTRIBUTION OF TROPICAL EEL GENUS Anguilla IN INDONESIAN WATER BASED ON SEMI-MULTIPLEX PCR Abstract ... 20
Introduction ... 20
Material and Methods ... 23
Result ... 25
Discussion ... 28
IV. A NEW MOLECULAR PHYLOGENY AND GENETIC DIVERSITY OF FRESHWATER EEL GENUS Anguilla IN INDONESIAN WATER BASED ON MITOCHONDRIAL GENES Abstract ... 35
Introduction ... 35
Material and Methods ... 38
Result ... 41
Discussion ... 52
V. POPULATION GENETIC STRUCTURE OF TROPICAL EEL Anguilla bicolor IN INDONESIAN WATER Abstract ... 55
Introduction ... 55
Material and Methods ... 58
Result ... 62
Discussion ... 65
VI. GENERAL DISCUSSION ... 67
REFERENCES ... 71
APPENDIX ... 76
LIST OF TABLES
Page
1. Position of nine species-specific primers for semi-multiplex PCR on
cytochrome b and 16s rRNA genes respectively. Two positions for
forward primers (FCYT-EEL and F16S-EEL) and seven positions for
reverse primers ... 15
2. Species-specific primer sequences for semi- multiplex PCR, and PCR product lengths expected for the seven Anguilla species and subspecies ... 16
3. Identification eel by morphology (only 796 eels were classified into the four groupa) and semi-multiplex-PCR (1112 eels were identified among 1115 eels) ... 18
4. List of sampling locations of tropical eels in Indonesia ... 25
5. List specimens, sampling location and date in this study ... 39
6. Matrix nucleotide substitution based on HKY+G+I methods ... 42
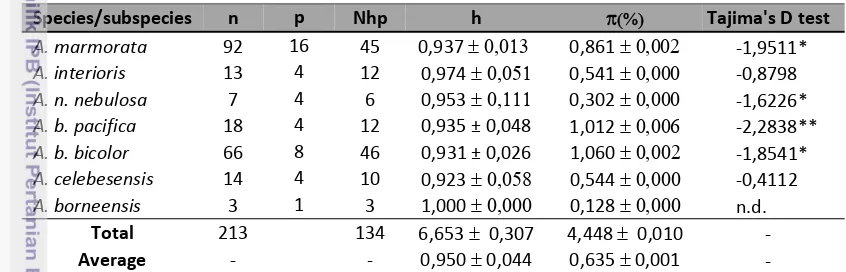
7. Genetic diversity and neutral test of species/sub species of tropical eel genus Anguilla from Indonesia waters ... 43
8. Nucleotide diagnostic of each species and clade on the species ... 47
9. Matrix genetic distance between all of species-subspecies on genus Anguilla. Grey highlights one is species-subspecies during this study ... 48
10. Genetic distance within species and sub species ... 48
11. Intraspecific genetic differentiation measured within A. bicolor species ... 51
12. Mutation on nucleotide sequence (above) and amino acid (below) between A. b. bicolor and A. b. pacifica ... 51
13. Intraspecific genetic differentiation measured within A. marmorata species ... 52
14. List of specimen used for microsatellite analyses in the present study .... 59
15. List locus that using during this study ... 61
16. Genetic variability of population parameters at 7 microsatellite loci on tropical eel in Indonesia water ... 63
17. Genetic divergence (Fst) (above) and genetic distance (below) among population during present study ... 64
LIST OF FIGURES
Page
18. World production of eels (FAO, 2009) ... 3
19. Research framework ... 9
20. Measurement morphology character of specimen ... 15
21. Identification species and subspecies of Anguilla by semi-multiplex PCR
samples ... 18
22. Map of the stations around Indonesia Sea where samples have been
collected for this study ... 25
23. Distribution of the seven species and subspecies of freshwater eels
which were found around Indonesia during this study ... 28
24. The sampling location of genus Anguilla in the origin region Indonesia
water ... 41
25. Neighbor Joining (NJ) phylogentic tree based on 1042 bp of
mitochondrial DNA cyt b gene fragmen under Kimura 2-parameter model,
with Serrivomer sector, Synaphobranchus kaupi and Conger myriaster as
outgroup (up to 75%) ... 46
26. Neighbor Joining (NJ) phylogentic tree based on 1042 bp of
mitochondrial DNA cyt b gene fragmen under Kimura 2-parameter model,
with Synaphobranchus kaupi as outgroup (up to 75%) ... 47
27. Neighbor Joining (NJ) phylogentic tree of all sequence in this study
based on 1042 bp of mitochondrial DNA cyt b gene fragmen under
Kimura 2-parameter model (up 75%) ... 51
28. Dendogram of genetic distance of Indonesian tropical eel based on cyt b
sequence ... 56
v
LIST OF APPENDIX
Page
1. Sampling Location during this Study ... 76
2. Number haplotype for the cytochrome b gene among all specimen on
present study ... 77
3. Sequencing Alignment for Cytochrome b gene among all haplotype of
Anopicall eel Anguilla spp ... 79
4. Frequency allele for each locus, each localities for all specimen ... 83
LIST OF TABLES
Page
1. Position of nine species-specific primers for semi-multiplex PCR on
cytochrome b and 16s rRNA genes respectively. Two positions for
forward primers (FCYT-EEL and F16S-EEL) and seven positions fo
reverse primers ...
2. Species-specific primer sequences for semi- multiplex PCR, and PCR r
... 15
... 16
into the
18
.... 25
. 39
... 61
64 product lengths expected for the seven Anguilla species and
subspecies ...
3. Identification eel by morphology (only 796 eels were classified
four groupa) and semi-multiplex-PCR (1112 eels were identified among
1115 eels) ...
4. List of sampling locations of tropical eels in Indonesia ...
5. List specimens, sampling location and date in this study ...
6. Matrix nucleotide substitution based on HKY+G+I methods ... 42
7. Genetic diversity and neutral test of species/sub species of tropical eel
genus Anguilla from Indonesia waters ... 43
8. Nucleotide diagnostic of each species and clade on the species ... 47
9. Matrix genetic distance between all of species-subspecies on genus
Anguilla. Grey highlights one is species-subspecies during this study ... 48
10. Genetic distance within species and sub species ... 48
11. Intraspecific genetic differentiation measured within A. bicolor species ... 51
12. Mutation on nucleotide sequence (above) and amino acid (below)
between A. b. bicolor and A. b. pacifica ... 51
13. Intraspecific genetic differentiation measured within A. marmorata
species ... 52
14. List of specimen used for microsatellite analyses in the present study .... 59
15. List locus that using during this study ...
16. Genetic variability of population parameters at 7 microsatellite loci on
tropical eel in Indonesia water ... 63
17. Genetic divergence (Fst) (above) and genetic distance (below) among
population during present study ...
LIST OF FIGURES
Page
1. World production of eels (FAO, 2009) ... 3
2. Research framework ... 9
3. Measurement morphology character of specimen ... 15
4. Identification species and subspecies of Anguilla by semi-multiplex PCR
25
1
del,
... 51
t b samples ... 18
5. Map of the stations around Indonesia Sea where samples have been
collected for this study ...
6. Distribution of the seven species and subspecies of freshwater eels
which were found around Indonesia during this study ... 28
7. The sampling location of genus Anguilla in the origin region Indonesia
water ... 4
8. Neighbor Joining (NJ) phylogentic tree based on 1042 bp of
mitochondrial DNA cyt b gene fragmen under Kimura 2-parameter model,
with Serrivomer sector, Synaphobranchus kaupi and Conger myriaster as
outgroup (up to 75%) ... 46
9. Neighbor Joining (NJ) phylogentic tree based on 1042 bp of
mitochondrial DNA cyt b gene fragmen under Kimura 2-parameter mo
with Synaphobranchus kaupi as outgroup (up to 75%) ... 47
10. Neighbor Joining (NJ) phylogentic tree of all sequence in this study
based on 1042 bp of mitochondrial DNA cyt b gene fragmen under
Kimura 2-parameter model (up 75%) ...
11. Dendogram of genetic distance of Indonesian tropical eel based on cy
sequence ... 56
I. GENERAL INTRODUCTION
Background
The freshwater eels (Anguilla spp: Anguillidae) are a well known for their
catadromous life histories. They migrate between freshwater and marine
environment. These species are breeding far offshore after a migration of
thousands kilometers from their growth habitats in freshwater and estuarine to
their spawning area in oceanic waters (Ege 1939; Tesch 1977). Most of the
temperate species spawn within a narrow tropical area. Furthermore, the eels
larvae, known as leptocephali, passively swim to their growth habitat along
subtropical currents (Tesch 1977; Tsukamoto 1992).
The anguillid leptocephalus is one of the most distinctive larvae of
Anguilliform fishes and has an olive leaf-like shape, with a depth of about one
fifth of total length (TL), no melanophores, transparent body, relatively few
myomeres and high water content contribute to the buoyancy, which would be
advantageous for passive transport by ocean current (Tesch 1977; Mochioka
2003). Lepthocephalus will be metamorphose, their morphological and
physiological change before into glass eel, which occurs before entering
freshwater. Eel larvae undergo marked changes the somatic structure from the
leaf-like shape of the lepthocephali to the adult-like shape of the glass eel. Glass
eel is a developmental stage from the end of metamorphosis to the beginning of
pigmentation, following this stage the young eels are called “elver” (Tesch 1977;
Mochioka 2003). The next development stage is the yellow eel or sexually
immature adult stage. Those inhabit in estuarine until freshwater environment,
which tend to live longer and attain much larger sizes (Tesch 1977). Further
freshwater eels will continue to the sexually mature stage are called silver eel. In
this stage, they begin downstream migration from the growth habitat, freshwater
to the open oceanic, after transformation from yellow-phase to silver-phase eels.
The silver eel spent their entire growth phase in the marine environment and
apparently they never entered freshwater (Tesch 1977; Aoyama et al. 2003a).
Ege (1939) divided the Anguilla species into four groups, based upon
color, body proportions, dentition, and meristic characters: First group, variegated
species with broad undivided maxillary and mandibular bands of teeth;
A. celebesensis Kaup 1856; A. interioris Whitley 1938; and A. megastoma Kaup
1856; second group, variegated species with a toothless longitudinal groove in
the maxillary and mandibular bands of teeth; A. nebulosa (A. n. labiata Peters
1852; A. n. nebulosa McClelland 1844); A. marmorata Quoy and Gaimard 1824;
A. reinhardti Steindachner 1867; and A. ancestralis Ege 1939; third group,
species without variegated markings and with a long dorsal fin; A. anguilla
Linnaeus 1758; A. rostrata Lesueur 1817; A. mossambica Peters 1852;
A. borneensis Popta 1924; A. japonica Temminck and Schlegel 1846; and
A. dieffenbachii Gray 1842; and fourth group, species without variegated
markings and with a short dorsal fin; A. bicolor (A. b. bicolor McClelland 1844;
A. b. pacifica Schmidt 1928); A. australis (A. a. australis Richardson 1841;
A. a. schmidti Phillips 1925); and A. obscura Gunther 1871. Castle and
Williamson (1974) postulated that A. ancestralis is an invalid species, based on
similarities with juvenile A. celebesensis. Currently, after scientist discovered a
new species A. luzonensis in Philippines water at 2009, accepted number of
species in the genus Anguilla is 16 species (Watanabe et al. 2009).
At the population level, most of the freshwater eel showed randomly
mating panmictic structure on the whole species or sub-species distribution
(Avice et al. 1986). Panmixia is realized on the unique species or subspecies
spawning area where all individuals are potential partners. There are no mating
restrictions, neither genetics or behavioral and all recombination is possible
(Avise et al. 1986).
Apart from their unique life history, eels are also popular as a commercial
important food, because of good nutritional value, with protein and fat content of
65% and 28% respectively (Hainsbroek 2007). Among the many popular eel
dishes consumed around the world, kabayaki (marinated grilled eel) is a national
dish in Japan, while smoked eel is favored in Europe and North America, and eel
larvae are eaten as appetizers in Spain (Ringuet 2002).
The international market for cultured eels exceeds 200,000 ton in year
2000. Based on FISHSTAT (FAO 2009) data, total production of eels rose from
17,750 ton in 1950 year to 284,274 ton in 2007 year. In Japan, the Japanese eel
(Anguilla japonica) has long been esteemed as an important food fish, as much
as 130,000 tons of eels are consumed per year followed by China, Korea,
America and some European countries, like Denmark, France, Italy, Belgium and
Germany. However most of this production is based on catching wild of adults
and rearing of wild-caught juvenile “glass eels”. The catching activity of glass eel
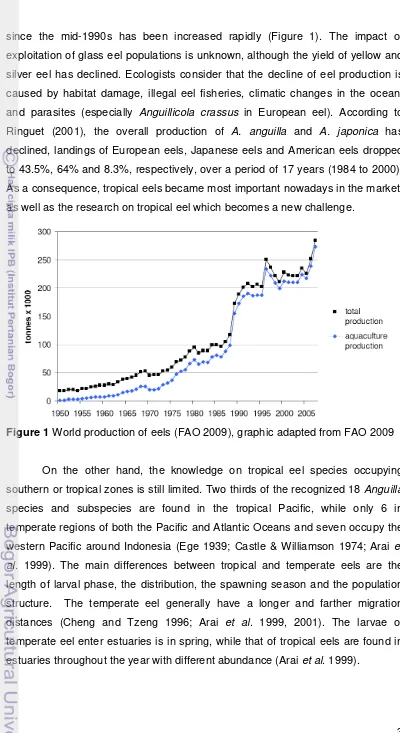
since the mid-1990s has been increased rapidly (Figure 1). The impact of
exploitation of glass eel populations is unknown, although the yield of yellow and
silver eel has declined. Ecologists consider that the decline of eel production is
caused by habitat damage, illegal eel fisheries, climatic changes in the ocean,
and parasites (especially Anguillicola crassus in European eel). According to
Ringuet (2001), the overall production of A. anguilla and A. japonica has
declined, landings of European eels, Japanese eels and American eels dropped
to 43.5%, 64% and 8.3%, respectively, over a period of 17 years (1984 to 2000).
As a consequence, tropical eels became most important nowadays in the market,
as well as the research on tropical eel which becomes a new challenge.
Figure 1 World production of eels (FAO 2009), graphic adapted from FAO 2009
On the other hand, the knowledge on tropical eel species occupying
southern or tropical zones is still limited. Two thirds of the recognized 18 Anguilla
species and subspecies are found in the tropical Pacific, while only 6 in
temperate regions of both the Pacific and Atlantic Oceans and seven occupy the
western Pacific around Indonesia (Ege 1939; Castle & Williamson 1974; Arai et
al. 1999). The main differences between tropical and temperate eels are the
length of larval phase, the distribution, the spawning season and the population
structure. The temperate eel generally have a longer and farther migration
distances (Cheng and Tzeng 1996; Arai et al. 1999, 2001). The larvae of
temperate eel enter estuaries is in spring, while that of tropical eels are found in
estuaries throughout the year with different abundance (Arai et al. 1999).
Most studies of population genetic structure of eels have focused just on
temperate species in the northern hemisphere and few have examined tropical
species. Some studies on population genetic structure of tropical eel have been
conducted such as; A. marmorata by Ishikawa et al. (2004), Tseng (2012) and
Minegishi et al. (2008); A. bicolor by Minegishi et al. (2012) and A. reinhardtii by
Shen and Tzeng (2007, 2012).
To uncover the existence of tropical eel, several studies have been
conducted through the collaboration between the Japanese institution Ocean
Research Institute and research institutes of Indonesia (LIPI) to investigate the
distribution of tropical eels , using the research vessels KM Baruna Jaya VII and
RV Haruko Maru in the period 1998-2003. Both research vessels collected
lepothocephali around the Indonesian sea: A. borneensis caught in the Celebes
Sea, A. b. bicolor caught in the Mentawai Islands and A. celebesensis in the
Tomini bay (Arai et al. 1999; Wouthuyzen et al. 2009; Aoyama et al. 2007;
Setiawan et al. 2001; Miller 2003).
Beside looking for the spawning areas of these eels, this expedition
confirmed also some observations quite interesting for taxonomy. According to
Watanabe et al. (2004a), the tropical eels show geographic distribution and
morphological characters heavily overlapping. The comprehensive identification
of eels proposed by Ege (1939), divided genus Anguilla into 15 species, three of
which were subdivided into two subspecies by using morphological character.
But when the geographic distribution of each species is plotted on a map, several
species have overlapping geographic range. They have also overlapping range
of morphological character. While Ege's taxonomy has long been accepted since
its publication (because most of the first freshwater eel studied came from
temperate region and are not geographically overlapping), some doubts were
expressed (Watanabe et al. 2004a).
That means the identification of eels depends on the location of collection.
Watanabe et al. (2004a) deduced that if only morphological characters were used
for identification, the freshwater eels could be classified into only four groups.
This can be also a problem for the freshwater eels that have been transported
around the world in recent years for aquaculture.
Since morphological studies are no more sufficient for freshwater eels
species identification, the molecular genetics approaches become a new
challenge to develop technically and theoretically nowadays. Molecular genetics
have been used to evaluate certain taxa, for identification, evolution purposes
and phylogenies reconstructions (Freeland 2005). The application of molecular
genetics to confirm identification of freshwater eel have been conducted in
several research (Aoyama et al. 1999, 2001; Watanabe et al. 2004b, 2008;
Sezaki et al. 2005; Itoi et al. 2005; Gagnaire et al. 2007 and Trautner et al. 2006),
most of them apply RFLP, RAPD and Real Time-PCR methods with 16S rRNA
and Cyt b gene as marker.
Multiplex PCR is a variant of PCR permitting simultaneous amplification of
several targets in one reaction by using more than one pair of primers. For taxa
identification purpose, the multiplex PCR produce amplicons on varying sizes that
are specific to different DNA sequences (Rompler 2006). Multiplex PCR assay
that using several species-specific primers enable to identify several species in a
simple, quick, low cost, sensitive, and highly reliable method (Catanese et al.
2010)
The molecular genetics techniques also have been used for new
hypotheses of phylogeny and evolution of the genus Anguilla (Tagliavini et al.
1996; Aoyama and Tsukamoto 1997; Lin et al. 2001; Bastrop et al. 2000;
Aoyama et al. 2001; Inoue et al. 2001 and Minegishi et al. 2005). Molecular
phylogenetic studies upon all species of genus Anguilla have been conducted by
Lin et al. (2001) who examined mitochondrial 12SrRNA and cytochrome bgenes,
and Aoyama et al. (2001) examined 16SrRNA and cytochrome b genes. Both
studies presented almost the same topology defining species groups and their
geographic distribution. However these contributions diverged on the position of
basal species. Aoyama et al. (2001) concluded that A.borneensis was most likely
the basal species of the genus while Lin et al. (2001) suggested that the ancestor
species are A. marmorata and A.nebulosa.In recent year, Minegishi et al (2005)
produced a new phylogenetic analysis based on the complete mitochondrial DNA
(mtDNA) sequence of all species of genus Anguilla. This phylogenetic analysis
showed a better statistical support that mean more sure analysis than previous
studied. Minegishi et al. (2005) suggest A. mossambica to be most basal species
using Baysian analysis, but based on MP (maximum parsimony) analysis,
similarly with Aoyama et al. (2001), Minegishi et al. (2005) A. borneensis appears
as basal species. Moreover, because the tropical and Indo-Pacific zones have a
highest species diversity, these authors said that the Indonesia waters are the
“origin of the eels” and “Indonesia is homeland of eels”.
A. bicolor and A. marmorata are tropical eels that have been widely
analyzed due to their exceptional large distribution (nearly 20,000 km east-west
in Indopacific oceans). This wide distribution is mostly in sympatry from the
eastern coasts of Africa through the seas around Indonesia to New Guinea in the
Pacific Ocean. They are strongly suspected to have several spawning areas.
According to Minegishi et al. (2008), A. marmorata is not taxonomically divided
into subspecies because of its morphological stability, but molecular studies have
demonstrated its structure into four differentiated populations: North Pacific,
South Pacific, Indian Ocean, and Mariana (Minegishi et al. 2008; Gagnaire et al.
2009). The shortfined eel, A. bicolor, of high abundance, is considered to be
structured into two subspecies A. b. bicolor in Indian Ocean especially at the west
of Indonesia and A. b. pacifica in Pacific Ocean (Ege 1939; Minegishi et al.
2012). However, the population structure and evolutionary history of A. bicolor
needs to be investigated in detail especially in Indonesia. These fish have high
economic value, and are believed to be the best candidate eels to replace
Japanese eel (A. japonica) and European eel (A. anguilla) which have been
decline for food, including fish farm growth.
Indonesia is an archipelagic country that has a long coastline of 91,000
km and 71,480 islands. The western part of Indonesia is connected with Indian
Ocean and the eastern was one with the Pacific Ocean, making Indonesia an
important biogeographic crossroad. Until now, there are seven recognized
species that occupy Indonesia waters: A. bicolor (two subspecies: A. b. bicolor
and A. b. pacifica), A. marmorata, A. celebesensis, A. borneensis, A. interioris, A.
obscura and A. nebulosa (subspecies: A. n. nebulosa) (Ege 1939; Castle &
Williamson 1974; Tsukamoto & Aoyama 1998 and Sugeha et al. 2008). But
information about distribution, evolution, phylogenetic relationship and structure
population still are limited and without details.
Questions to solve
This study was intended to solve some questions:
a. The distribution of seven eel species living in Indonesia is not really known.
This project aimed first at establishing quick methods of identification of
tropical eels. This is partly due to the difficulty to determine the species just
using morphological character. From here we begin to get a clear description
of the distribution of the species that live in Indonesian water. while it is quite
b. The genetic relationships among Anguillide in Indonesian water can be used
generate a new phylogenetic tree which can help to understand the evolution
of this genus in tropical areas especially in Indonesian water.
c. Indonesia is an important biogeographic crossroad, at the contact between
Indian and Pacific ichthyofaunas. Concerning the two widespread eel
species (A. marmorata and A. bicolor), Indonesia harbors a part of the Indian
and a part of the Pacific populations or subspecies, but nearly nothing is
known on the exact distribution of each lineage in Indonesian rivers.
Biogeography of both species and populations genetics of A. bicolor will be
used to understand the population structure and to know the gene flow
pattern among Anguillidae populations in the Indonesian waters.
Framework
The decreasing of temperate eel populations in the subtropical zone have
encouraged biologists to help conservation and aquaculture of this group. As a
result, research on tropical eel are become a new challenge especially in
Indonesian waters. Many scientists consider Indonesia as ”homeland and origin
of eels”, however, knowledge about distribution and biological traits of eel in
Indonesian waters is still limited. Understanding biological aspects and population
is an important point in the development of sustainable aquaculture.
In sustainable aquaculture, phylogeography and genetic population studies
are two first crucial steps to manage the fisheries resources. Such studies
should provide valuable information about their status, population fragmentation
and dispersal pattern. To analyze phylogeography of eels, we should begin by
their distribution map and phylogenetic relationships. Therefore samples
collection and identification become an important point. Recently many biologists
agree that identification of eels is more appropriate using molecular biology
approach. Semi-multiplex PCR is one method that provides genetic information to
identification of target species, by using the species-specific primers.
Semi-multiplex PCR assays can identify several species in a simple, quick, low cost,
sensitive and highly reliable way on one step PCR. Phylogenetic relationships
can be analyzed based on the information resulting from mtDNA sequencing.
The PCR and DNA sequencing constitute easy and fast methods, furthermore,
computer software provide convenient analytical methods to infer phylogenetic
relationships and evolution and some statistical calculation.
The analysis of polymorphic microsatellites will provide inter and intra
populations information structure. This population analysis should be conducted
on widespread species.
As a summary, the outputs of this research are (i) a quick identification
method of ells species living in Indonesian waters; (ii) distribution of eels species,
furthermore (iii) the information on systematics, phylogenetics and populations
genetics structure, which will provide data for conservation and aquaculture
strategy. The framework of this study is summarized in the Fig. 2.
Phylogeography tropical eels genus Anguilla in Indonesian waters
Distribution and species composition of Anguilla spp in Indonesian water
Phylogenetic tree tropical eel in Indonesian waters
Population genetic structure of tropical eel: Anguilla bicolor in Indonesian waters
CYT b and 16SrRNA genes to construct phylogenetic
relationships among populations and species Morphometric and
Semi-Multiplex PCR for identification
Polymorphic microsatellites to determine the population genetic structure of
A. bicolor
Quick identification methods, dispersal and distribution, systematic and phylogenetic and population genetic structure of tropical eels in
Indonesian water
Population connectivity and conservation-aquaculture strategy of tropical eel in
Indonesian waters
Figure 2. Research framework
II. A Novel Semi-Multiplex PCR Assay for Identification of
Tropical Eel Genus
Anguilla
in Indonesian Water
Abstract
A
o
ne step semi-multiplex PCR is proposed for distinguishing seven species andsubspecies of tropical eels including Anguilla bicolor bicolor, A. bicolor pacifica, A.
marmorata, A. interioris, A. celebesensis, A. borneensis, and A. nebulosa nebulosa in
Indonesian waters. Seven pairs of species-specific primers, including two forward and
seven reverse primer sequences, were designed after the alignment of complete
mitochondrial cytochrome b (1140 bp) and 16S rRNA (1120 bp) genes. All
species-specific primer pairs are included in one PCR, but only one pair of them can amplify a
specific fragment from the template DNA that is analyzed. The semi-multiplex PCR
amplified a fragment of 230 bp for A. b. bicolor, 372 bp for A. n. nebulosa, 450 bp for A.
borneensis, 620 bp for A. marmorata, 670 bp for A. b. pacifica, 720 bp for A.
celebesensis, and 795 bp for A. interioris, which are then separated by DNA agarose gel
electrophoresis.
KEY WORDS: semi-multiplex PCR, Anguilla, tropical eels, molecular identification, species-subspecies specific primer
Introduction
Order Anguilliformes contains 400 genera and 800 species, most of them
lives in the oceans and only genus Anguilla migrates to the freshwater for growth
Nelson (2006). Genus Anguilla is composed of freshwater eel having a
catadromous life history characterized by spawning in ocean waters and by a
migration of the larvae back to the parents growing habitats in freshwater or
estuarine areas. The three temperate species spawn in remote tropical waters
after a long adult migration. Their larvae, called leptocephali are passively
returning to their growth habitat with the influence of subtropical currents, and
perform long migration distance (Tesch 1977; Tsukamoto 1992). Oppositely, the
tropical eels have shorter migration distance for both adults and leptocephali than
those of the temperate species (Arai et al. 1999; Wouthuyzen et al. 2009 and
Aoyama 2009)
A first comprehensive identification of the genus Anguilla was proposed by
Kaup (1856), who recognized 45 species. In 1870, Gunther reduced this number
to 23 species. Lastly, a revision of the genus was done by Ege (1939) who
divided the genus Anguilla into 16 species, three of which were subdivided into
two subspecies. Based on morphological characters, the systematic organization
of Ege (1939) have long been widely accepted by many biologists. However,
Watanabe et al. (2004a) recently found that the morphological character
described by Ege (1939) were not sufficient to classify all species of this genus
without including the information on the geographic distribution of the specimens,
which he used as a taxonomic character. Watanabe et al. (2004a) considered
that the Ege’s (1939) key for species identification is partly invalid because many
morphological characters used are overlapping in most species. Species
recognition becomes especially important for tropical eels because nowadays
they are commercially transported around the world for food (frozen) and
aquaculture (a live) purposes. Ege's (1939) key is considered as insufficient
especially in tropical areas, where geographic and morphological characters of
eels are heavily overlapping and where scientific data are still scarce.
Indonesia is a wide equatorial characterized by archipelagos composed of
around 71,480 islands and coastline of around 91,000. Biogeographically,
Indonesia is at the crossroad of the Indian and Pacific Oceans, the western part
of Indonesia is connected with Indian Ocean and the eastern is connected with
the Pacific Ocean. Two thirds of the recognized 18 Anguilla species and
subspecies are found in the tropical Pacific and seven species and subspecies of
tropical eels range around Indonesia (Ege 1939; Castle & Williamson 1974, Arai
et al. 1999). Those are A. bicolor (two subspecies: A. b. bicolor and A. b.
pacifica), A. marmorata, A. celebesensis, A. borneensis, A. interioris, A. obscura
and A. nebulosa (subspecies: A. n. nebulosa). As a result, the position of
Indonesia appears to be center of origin the diversity of eel and is strategic in the
knowledge of their evolution.
Several methods for species identification have been used on fish like
conventional morphology and electrophoresis, immunoassay, liquid
chromatography or molecular genetics assay (O’Reilly and Wright, 1995).
Concerning eels, after demonstration that morphological characters were not
sufficient to classify all species, molecular genetics have been recommended
(Watanabe et al. 2004a). Most of the genetic approaches to species identification
are based on the amplification of a partial sequence of mitochondria (mtDNA).
The mtDNA, of maternal inheritance, shows no recombination, so that its
sequences are more conservative. The gene 16S rRNA, relatively well
conserved, is considered as a good marker for genera and species identification
(Aoyama et al. 2000, 2001; Watanabe et al. 2004b, 2005; Gagnaire et al. 2007).
However, it is not polymorphic enough between subspecies (Watanabe 2003).
Moreover, it is important to develop the markers that could distinguish
subspecies. One of the other genes used is cytochrome b (Cytb). This functional
gene is positioned between tRNAGlu and tRNAThr genes. Many investigations on
Cytb focus on inheritance and evolution (Freenlad 2005). Several studies have
used Cytb as a marker for identification of subspecies (Jain et al. 2008; Hyde et
al. 2005).
Recently, several simple PCR techniques have been used to distinguish A.
japonica and A. anguilla (Sezaki et al. 2005), A. interioris and A. celebesensis
(Aoyama et al. 2000) and to distinguish A. anguilla and A. rostrata (Trautner
2006). Application of Real-Time PCR technique allowed identification of A.
japonica leptocephali (Watanabe et al. 2004b), using single nucleotide
polymorphism (SNP) (Itoi et al. 2005), Random Amplified Polymorphic DNA
(RAPD) (Kim et al. 2009; Lehmann 2000), Restriction Fragment Length
Polymorphism (RFLP) (Lin et al. 2001), most of these researches dealing with
temperate area.
Rapid molecular identification of tropical eel began with Gagnaire et al.
(2007), who developed semi-multiplex PCR and RFLP to identify four eel species
in Indian Ocean. Multiplex PCR is a variant of PCR enabling simultaneous
amplification of several targets in one reaction by using more than one pair of
primers. The multiplex PCR produces amplicons on varying sizes that are
specific to different DNA sequences (Rompler 2006). By using the
species-specific primers in multiplex-PCR assays, the identification of several species in a
simple, quick, low cost, sensitive, and highly reliable amplification is possible
(Catanese et al. 2010).
In the present study we developed a method derived from multiplex PCR
assay. In this method, the PCR is based on two “universal” forward Anguilla
primers (and so, called semi-multiplex) and seven specific reverse primers, one
for each eel species or subspecies know from Indonesian waters. The method is
based on the complete sequence of cytochrome b and 16S rRNA extracted from
the specimens that was collected around Indonesia waters.
Material and Methods
Specimens
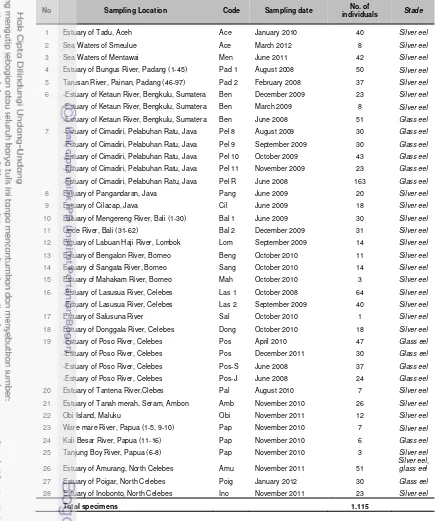
The 1115 specimens examined in this study were collected around the
Indonesian waters, covering all the geographic distribution of Anguilla species
that were expected to occur in the country (see Appendix 1). Around 800
specimens are silver eels while the others are glass eels. Samples collection
was conducted in rivers estuaries along the coasts of Indian Ocean, Pacific
Ocean and around Arafura and Celebes Seas. The specimens were collected
from 2008 to 2012. Species assignation was preliminary performed by using
available morphological keys (Watanabe et al. 2004a and Reveilac et al. 2007).
The first morphological characters, which are measurements in this study, a
quantitative one as follows: the total length (LT), the dorsal fin length (LD), the
anal fin length (LA). These measurements were used to calculate the distance
between the origin of the dorsal and anal fins (DA) using the formulation
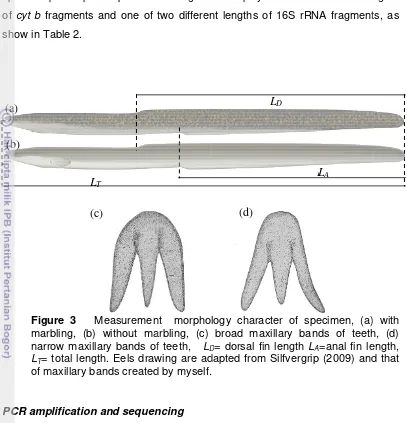
DA=100(LD – LA)LT-1 (Reveillac et al. 2007) (Fig.3). This character determined
whether an individual was short-fin (FS) or long fin (FL). The accuracy of the
measurement is 0.01 mm by using the digital caliper. The second measurement
of morphological character was qualitative parameters that are presence or
absence of marbling and breadth of maxillary bands. According to Watanabe et
al. (2004a) genus Anguilla can be divided into four groups based on three
characters: presence or absence of marbling, wide and narrow maxillary band of
teeth and origin of the dorsal fin; group 1, groups long dorsal fin with marbling
skin and broad maxillary bands of teeth; group 2, groups long dorsal fin with
marbling skin and narrow maxillary bands of teeth; group 3, groups long dorsal fin
and no marbling and group 4, groups short dorsal fin without marbling skin.
Tissues from anal fin, which were immediately stored in 95% ethanol, were used
for genetic analysis.
Design of PCR primer
Nine semi-multiplex PCR primers were designed from sequence
alignment performed on the cytochrome b (cyt b) and the 16S rRNA genes. The
Cyt b and 16S rRNA sequence dataset include sequence from GenBank (ref.
AP007236, AP007237, AP007238, AP007239, AP007241, AP007242,
AP007246) and 100 sequences obtained during the present study. Nine position
of original sets for semi-multiplex PCR primers are shown in Table 1. Each
species-sp
of cyt b fr
show in Ta
pecific prim
ragments a
able 2.
mer pairs wa
nd one of t
as designed
two differen
d to amplify
nt lengths o
one of five
of 16S rRN
different le A fragment ngths ts, as Figu mar narr LT=
of m
PCR amp
The fi
suitable a
PCR in a
the primer
volume of
(25mM), 1
primer (co
(5u/µl),
(b)
(a)
ure 3 M
bling, (b) w row maxillar
total length maxillary ban
plification a
irst step of
annealing te
Mastercycle
rs were mix
f 10 µl cont
1.25 µl dN
ommon pri
0.2 µl d
L
T(c)
easuremen without ma ry bands of h. Eels draw
nds created
and sequen
semi-multip
emperature
er gradient
xed in one P
taining 2 µl
TP (2mM),
mer) is 1
ddH2O an
nt morpho arbling, (c)
f teeth, LD
wing are ad d by myself.
ncing
plex PCR a
for all spe
(Eppendorf
PCR solutio
5x Green
0.5 µl eac
µl (10mM)
nd 1 µl
logy charac broad max
D= dorsal fin
dapted from
mplification
ecies-specif
f, Le Pecq,
on. The PC
GoTaq@ re
ch primer
), 0.05 µl
template
(d)
L
Dcter of spe xillary band n length LA
m Silfvergrip
n was to est
fic primers
France). In
R was carr
eaction buff
(10mM) ex
GoTaq@ D
DNA (
L
L
Aecimen, (a) ds of teeth
=anal fin le (2009) and
) with h, (d) ength,
d that
tablish the s
by using s
second ste
ried out in a
fer, 0.5 µl M
xcept FCYT
DNA polyme
Table 1 Position of nine species-specific primers for semi-multiplex PCR on cytochrome b and 16s rRNA genes respectively. Two positions for forward primers (FCYT-EEL and F16S-EEL) and seven positions for reverse primers. See Table 2 for nucleotide sequence of primers.
Semi-multiplex-PCR was carried out in a Thermal Cycler from Bio-Rad,
programmed to perform a denaturation step at 95oC for 5 min, followed by 35
cycles consisting of 45s at 95oC, 45s at 50oC (see Results) and 1 min at 72oC.
The final extension step (at 72oC) was 10 min. Five microliters of each PCR
product were loaded on a 1,5% agarose electrophoresis gel, stained with Cyber
Safe before electrophoresis at 100 volt for 90 min. The DNA band were observed
under Blue Light and photographed by Canon camera digital
Five individuals of each banding pattern were sequenced on cytochrome b
gene with the primer pair F-EEL-Cytb: 5’ CCA CCG TTG TAA TTC AAC 3’ and
R-EEL-Cytb: 5’ AAG CTA CTA GGC TTA TC 3’. To ensure the identification of
species, alignment was done by using Mega 5.0 (Kumar et al. 2008) and
compared with published sequences.
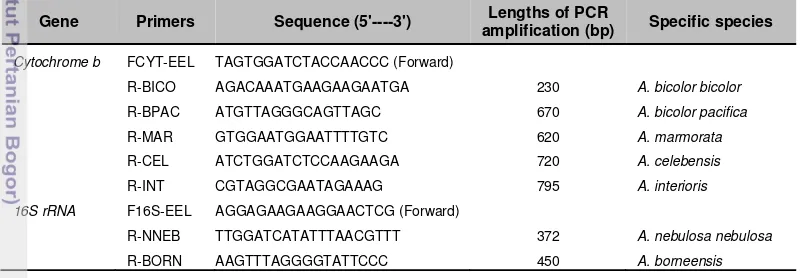
Table 2 Species-specific primer sequences for semi- multiplex PCR, and PCR product lengths expected for the seven Anguilla species and subspecies
Gene Primers Sequence (5'----3') Lengths of PCR
amplification (bp) Specific species
Cytochrome b FCYT-EEL TAGTGGATCTACCAACCC (Forward)
R-BICO AGACAAATGAAGAAGAATGA 230 A. bicolor bicolor
R-BPAC ATGTTAGGGCAGTTAGC 670 A. bicolor pacifica
R-MAR GTGGAATGGAATTTTGTC 620 A. marmorata
R-CEL ATCTGGATCTCCAAGAAGA 720 A. celebensis
R-INT CGTAGGCGAATAGAAAG 795 A. interioris
16S rRNA F16S-EEL AGGAGAAGAAGGAACTCG (Forward)
R-NNEB TTGGATCATATTTAACGTTT 372 A. nebulosa nebulosa
R-BORN AAGTTTAGGGGTATTCCC 450 A. borneensis
Result
Nine species-specific primers have been designed after the alignment of
complete cytochrome b and 16S rRNA genes, including two forward primers
(FEEL-CYT and FEEL-16S) and seven reverse primers (BIC0, BPAC,
R-MAR, R-CEL, R-INT, R-NNEB and R-BORN) (see Table 2). The seven
species-specific fragments were successfully amplified at 50oC annealing temperature.
Five species and subspecies of tropical eel were distinguished by cytochrome b
gene with one forward and five species-specific reverse primers are A. b. bicolor,
A. b. pacifica, A. marmorata, A. celebesensis and A. interioris. Two species were
distinguished by 16S rRNA gene with one forward and two species-specific
reverse primers are A. n. nebulosa and A. borneensis. For the series analyses,
the whole 9 primers were added in the mix amplifiying their corresponding DNA
fragm
semi
beca
ampl
ments in on
i-multiplex
ause of the
lification fra
e step PCR
PCR assay
eir bad qua
agments.
R. A total of
y. Only 3 o
ality DNA.
f 1115 sam
out of 1115
Figure 4 s
ples have b
5 sample co
shows sev
been amplif
ould not be
ven differen
fied by this
e amplified
nt sizes of
M 1 2 3 4 5 MM 6 7 8 9 10 11 12 133 14 15 16 177 M 18 19 200 21 22 M 23 24 25 26 27 M
395 F T b m u T Unsp deter minu F in or also three indiv were unsp
but o
silve beca (Moc spec ident Morp pairs
Figure 4. Iden The orde of th b. pacifica, 11 marmorata, 23 unexpected ba
The sizes o
pecific ban
rmination.
utes migratio
Five individ
rder to conf
done for in
e specimen
vidual in line
e confirmed
pecific band
The quan
only 769 sp
r eel stage
ause the m
chioka 200
cimens is sh
tification of
phological a
s of taxa: A 5 bp
230 bp
ntification spec he sample is a
-12 are A. bor 3-25 and 27 a and.
of these se
nds are a
The differe
on of the PC
duals of eac
firm the exp
dividuals sh
ns showed
e 27 to Fig
d as A. int
s of 230 an
titative mea
pecimens po
e. The qua
orphologica
03). The fo
hown in Tab
all specim
analyses fa
A. celebese 230 bp
670
cies and subs as follow : 1-2 rneensis, 13-1 are A. interioris
even specie also produ ent fragme CR product ch banding pected iden howing une d unexpec
gure 4). One
terioris. A
nd 620 bp.
asurement
ossible to m
alitative me
al character
our morph
ble 3. The
ens (excep
iled to iden
ensis versu 0 bp
450 bp
pecies of Ang are A. n. nebu 17 are A.celeb s, M=100 bp l
es-specific f
uced, whic
nts sizes c
on a 1,5 %
pattern typ
ntification. S
expected ba
cted band
e of showe
As a result,
of DA was
measureme
asurements
rs in this s
ological gr
right side o
pt 3), based
tify glass e
us A. interi 720bp b
guilla by semi-ulosa, 3-7 are besensis, 18-2 adder, u
fragments a
ch does n
can be eas
% agarose e
pes were se
Sequencing
anding patte
among 1
ed DNA dam
, A. interio
applied on
ent by a qua
s are inapp
tage are no
rouping dis
of Table 3 i
d on semi-m
eels and can
rioris, A. n. 620bp b
multiplex PCR A. b. bicolor, 22 and 26 are nspecific band
are given i
not impede
ily observe
lectrophore
equenced a
g and alignm
ern. In this s
115 exam
mage and t
oris can pro
n all 1115 s
alitative one
plicable on
ot clearly e
stinguishing s species-s multiplex P nnot disting nebulosa bp 795 b R samples. 8-10 are A. A.
ds, =
n Table 2.
e species
ed after 90
esis gel. nd aligned ment were study, only ined (see two others oduce two specimens,
e that only
glass eel
established
g the 769
subspecies
CR assay.
guish three
marmorata and A. b. bicolor versus A. b. pacifica. Only A. borneensis is
identifiable by morphological character only, as a “long fin” without marbling eel.
These results are in agreement with Watanabe (2003).
Table 3. Identification eel by morphology (only 796 eels were classified into the
four group) and semi-multiplex-PCR (1112 eels were identified among 1115 eels)
Group by morphology (n) Species (n) by multiplex PCR
− Long dorsal fin with marbling skin and broad maxillary bands of teeth (15)
: A. celebenesis (47), A. interioris (16)
− Long dorsal fin with marbling skin and narrow maxillary bands of teeth (428)
: A. marmorata (487), A. n. nebulosa (15)
− Long dorsal fin, without marbling skin (3) : A. borneensis (3)
− Short dorsal fin, without marbling skin (323) : A. b. bicolor (510), A. b. pacifica (34)
Discussion
Since morphological identification was not sufficient to determine Anguilla
species and subspecies, several molecular approaches have been proposed on
previous study. Most of the identification methods used simple PCR followed by
sequencing (Sezaki et al. 2005; Trautner 2006). However, sequencing is not
convenient for large samples because of its cost in time and money. Some of the
studies have used multiple loci such as RFLP-PCR (Aoyama et al. 2001, 2000;
Lin et al. 2001) and RAPD-PCR (Kim et al. 2009) with non species-specific
primers, which mean that many loci are needed for identification. Besides, the
number of species determined with each above-mentioned method is limited,
such as two species (Aoyama et al. 1999; Sezaki et al. 2005) or four species
(Kim et al. 2009).
The semi-multiplex method proposed here has demonstrated to be efficient
for identifying seven species and sub-species of tropical eel with only one step
PCR. By using this method, one could reduce the number of necessary
sequences while the results are very sure for each species determination (we
easily identified 1112 specimens). All of species were obtained in this study have
show overlapping morphology and distribution. Moreover for small specimens
(mainly glass eels), the molecular method appears as indispensable.
Shen et al. (2010) suggested several additional criteria which must be
taken into account when considering multiplex PCR assay 1) minimize primer
dimer association between all of the primers; 2) similarity of the annealing
temperature of each primer; 3) primer specificity; and 4) constraint the migration
of the amplicons in order to separate the DNA fragments in agarose gel
18
electrophoresis. In the present survey each primer is species or
subspecies-specific and the limited cases of unexpected (non-subspecies-specific) amplification never
reduced the liability of species determination. Banding pattern of A. b. pacifica
shows a weak specific fragment of 670 and a bigger un specific band of 230, but
this fragment pattern being stabile for all our A. b. pacifica specimens, there is no
difficulty to determine this species.
The length of amplified fragments was clearly distinguishable after
electrophoresis migration (no overlapping fragment). The primer structure was
checked in order to avoid inter-molecules interaction, which is an important
precaution with 9 primers simultaneously mixed in one step PCR. Optimization of
the semi-multiplex PCR mix consisted in designing an annealing temperature and
quantity of primer permitting was similar to amplification efficiency to pairs of
primers.
Semi-multiplex PCR methods has been introduced recently on eels by
Gagnaire et al. (2007) in order to rapidly determinate tree species of Anguilla: A.
marmorata, A. megastoma and A bicolor bicolor from West Indian Ocean.
Genetic distances between each species pairs in genus Anguilla are almost 0,02
until 0.05 based on 16S rRNA (Watanabe 2003). This study has established a
rapid method to distinguish even two subspecies A. b. bicolor and A. b. pacifica,
although these two subspecies have low genetic distance which is 0.0068 based
on the 16S rRNA
This method has proven to be the most simple, quicker, lower cost (no
acrilamide migration), specific/sensitive, and highly reliable way than the other
ones used before. This rapid method provides a useful tool for aquaculture,
global marketing, and academic-scientific research.
III. Distribution of Tropical Eel Genus
Anguilla
in
Indonesian Waters Based on Semi-multiplex PCR
Abstract
Tropical eels living in Indonesian waters are known to be composed of
several species, but their real listing together with their distribution ranges need
to be established. The main difficulties are the very high number of islands with
perennial rivers where these species are living during the growth phase of their
life cycle. It is difficult, sometimes impossible, to determine the species using
morphological characters, moreover on glass eels. In order to establish the
geographic distribution of tropical eels of the genus Anguilla in Indonesian
waters, a total 1115 specimens were collected between 2008 and 2012. Sample
collection was done in the growth habitats that are rivers and estuaries by
commercial nets of different categories according to the fish size. All samples
were identified genetically using the recently developed semi-multiplex PCR
method. Four species and subspecies was recognized with wide distribution:
Anguilla bicolor bicolor, A. b. pacifica, A. marmorata and A. interioris; two species
with limited distribution, close to endemism: A. celebesensis and A. borneensis
and one subspecies A. nebulosa nebulosa that is only spread in river flowing into
Indian Ocean.
Key words:Anguillaspp, semi-multiplex PCR, tropical eel, distribution range,
Indonesian waters
Introduction
The catadromous freshwater eels genus Anguilla is distributed nearly
world-wide except the South Atlantic and the Eastern Pacific oceans (Ege 1939).
Freshwater eels spawn in the offshore ocean. After hatching, their larvae migrate
to coastal areas as pelagic, floating and transparent (Mochioka 2003). Eel larvae,
called leptocephali, are transported passively by warm currents flowing at low
latitudes. When they approach the continental shelf, leptocephali metamorphose
into glass eels before settling in the continental waters (rivers and lakes) to grow
for years until changing into yellow eels or “elver” and then silver eels. The
dispersal of leptocephali not only drives the distribution of freshwater eel species
on continental areas, but also the phylogeography of the genus and its evolution
(Aoyama and Tsukamoto 1997).
After Johannes Schmidt succeeded collecting anguillid leptocephali in the
Sargasso Sea in 1922 (Schmidt 1922), he and his colleagues, through Carlsberg
Foundation’s Oceanographic Expedition, continued their efforts by searching for
the spawning areas of freshwater eels in the Indo-Pacific region where most of
the species of this genus are found. They successfully collected leptocephali in
the Indo-Pacific region during their expedition from 1928 to 1930 (Jespersen
1942). However, most of these leptocephali have overlapping morphological
characters, hampering exact identifications. Since then, the spawning areas of
the Indo-Pacific anguillid species have remained a mystery. As a result, the
studies of Indo-Pacific eels are still poorly understood as well as the exact
locations of the spawning areas, and their larval migrations and the recruitment
mechanisms.
To solve the problems in identifying anguillid leptocephali, genetic
approaches, mtDNA sequences or RFLP, has been successfully used (Aoyama
et al. 1999, 2001a, 2001b; Aoyama 2003; Watanabe et al. 2005). Since species
identification of anguillid leptocephali has been developed, projects aimed at
learning more about the spawning areas, larvae distribution and larval ecology of
anguillid in the Indo-Pacific region have been organized. The long scientific cruise
of the Baruna Jaya, in central Indonesia sea, around Sulawesi Island, from
2001-2002, successfully collected leptocephali of A. marmorata, A. bicolor pacifica and
A. interioris. This survey also collected leptocephali of A. celebesensis and A. borneensis allowing to deduce the spawning areas of these species (Aoyama et al. 2003, Wouthuyzen et al. 2009). In 2003 this cruise also collected young
leptocephali A. bicolor bicolor in west Sumatera, positioning a spawning area of
A. b. bicolor in this zone (Aoyama et al. 2007).
The three Indonesian endemic species spawning areas are also to be
discovered: leptocephali of A. interioris have been caught in western Sumatera
waters (Aoyama et al. 2007) and Sulawesi waters (Aoyama et al. 2003,
Wouthuyzen et al. 2009); leptocephali of A. celebesensis were recognized in
Tomini Bay Sulawesi Island and that of A. borneensis were found in Makasar
strait (Aoyama et al. 2003; Wouthuyzen et al. 2009).
According to the geographic range of each species, Aoyama et al. (2001)
and Lin et al. (2001) who studied genus Anguilla molecular phylogenetics based
on partial mtDNA, showed four major subgroups in the world, that are Oceanic,
Atlantic, Tropical-Pacific and Indo-Pacific lineages. However, the complete
mtDNA sequence of all species of genus Anguilla was determined recently by
Minegishi et al. (2005) which suggested the geographic structure of eel as follow:
two Atlantic species (A. rostrata and A. anguilla), two Oceanic species (A.
differenbachii and A. australis), and nine Indo-Pacific species (A. japonica, A.
reinhardtii, A. marmorata, A. nebulosa, A. bicolor, A. interioris, A. celebesensis,
A. megastoma and A. obscura). Seven species/subspecies of the Indo-Pacific
Anguilla occur in the western Pacific and eastern Indian Ocean around
Indonesian waters (Ege 1939; Castle & Williamson 1974). Because of this high
diversity, several scientists considered that “Indonesia is homeland of anguilid”.
Besides, Indonesia is also known as "the origin of anguillid" because
phylogenetic analyses indicated that the endemic tropical species A. borneensis
from eastern Borneo (Kalimantan) is one of the most basal species of the genus
(Aoyama et al. 2001; Minegishi et al. 2005).
Among the 19 existing eel species and sucspecies, 7 occupy the
Indonesian rivers. Half of them are endemic, limited to Indonesia (A. borneensis,
A. cebesensis, A. interioris), the others show larger range distribution (A.
obscura, A. nebulosa), sometimes established at the Indo-Pacific level (A. bicolor
and A. marmorata). However, the species range distribution of the 7 Indonesian
species is only an extrapolation of very limited records.
The widespread species A. bicolor and A. marmorata are of interest
because their distribution and abundance place them in central economic
position, and because their exceptional geographic distribution (over 18,000 km
east-west) and structure made them an important model for eel biology
understanding, encompassing probably several spawning areas. Short fin eels A.
bicolor are considered to be structured into two subspecies A. b. bicolor in Indian
Ocean, especially at the west of Indonesia and A. b. pacifica in Pacific Ocean, at
the east of Indonesia (Minegishi et al. 2012).The giant mottled eel A. marmorata
is not taxonomically divided into subspecies because of its morphological
stability, however molecular studies have demonstrated its structure into four
differentiated populations: North Pacific, South Pacific, Indian Ocean, and
Mariana (Minegishi et al. 2008; Gagnaire et al. 2009, 2011).
One of the most exigent parts of this study is sampling strategy. Indonesia
is an archipelago country with counts 17000 islands, among them several
thousand islands possibly hosting eels so a complete sampling is impossible. The
selection of representative sampling location is necessary.
Indonesian sea have a complex topography and connectivity between the
Pacific and Indian Oceans, so surface heat fluxes, thermo cline and variability in
thermocline waters and patterns of Indonesia current was influenced by Pacific
and Indian Ocean fluctuations. The eastern part of Indonesia waters influenced
by the Indonesian throughflow (ITF) current. ITF are water mass flow passing
through Indonesian waters from Pacific to Indian Ocean. This water mass flow
occurs as a result of the pressure difference between the two oceans. The water
mass drives upper thermocline water from the North Pacific through the western
route of the Makassar Strait directly exit through the Lombok Strait or flow
eastward into the Banda Sea and further joined with the south equatorial current
(SEC) (Wirtky 1973; Gardon 2005). The western Indonesian waters affected by
South Equatorial Counter Current (SECC), and this current going down the
Sumatran coast entry to southern coast of Java by South Java Current (SJC).
A new molecular identification method has been developed recently to
distinguish seven tropical eel inhabiting Indonesian waters, called semi-multiplex
PCR assay. By using multiple species-specific primers in one PCR reaction, the
identification of all Indonesian species is now simple, quick, low cost, sensitive,
and highly reliable (Fahmi et al. 2012).
In this study the semi-multiplex PCR methodology was used in order to
establish a first map distribution of species and subspecies of tropical eel
(Anguilla spp) that inhabit in Indonesian water.
Material and Methods
The 1115 specimens were collected in 28 locations around the
Indonesian waters, covering the whole geographic distribution of genus Anguilla
as known or expected in Indonesian waters (Appendix 1, Table 4). Specimens
collection was conducted in river estuaries along the coast of Indian Ocean,
Pacific Ocean and around Arafuru and Celebes Seas. The specimens were
collected from 2008 to 2011 by using traps, nets and fishing. Oceanographic
data of Indonesian seas (Gardon 2005) used for looking the movement of water
and the possible spread of eels.
All of specimens were identified using semi-multiplex PCR protocol
according to Fahmi et al (2012 (in press)). Nine species-specific primers were
added in one PCR reaction. The PCR was carried out in a total volume of 10 µl
containing 2 µl 5x Green GoTaq@ reaction buffer, 0.5 µl MgCl2 (25 mM), 1.25 µl
dNTP (2 mM), 0.5 µl each primer (10 mM) with 1 µl (10 mM), 0.05 µl GoTaq@
DNA polymerase (5 u/µl), 0.2 µl ddH2O and 1 µl template DNA (around 20 ng).
Semi-multiplex-PCR was carried out in a Bio-Rad Thermal Cycler, programmed
to perform a denaturation step at 95oC for 5 min, followed by 35 cycles with 45 s
at 95oC, 45 s at 50oC and 1 min at 72oC. The final extension step (at 72oC) lasted
10 min. Five microliters of each PCR product were loaded on a 1,5% agarose
electrophoresis gel, stained with Cyber Safe before electrophoresis and migrated
at 100 volts for 90 min. The DNA bands were observed under Blue Light and
photographed by a Canon camera digital.