i
ENDOPHYTIC FUNGI
OF WILD ZINGIBERACEAE FROM
MOUNT HALIMUN SALAK NATIONAL PARK
NICHO NURDEBYANDARU
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
2013
iii
STATEMENT
Hereby I declare this thesis entitled Endophytic Fungi of Wild Zingiberaceae from Mount Halimun Salak National Park is my research report that was carried out and written under guidance of my supervisory boards. This thesis has not been submitted in any form to any higher education institution except to Bogor Agricultural University (IPB). All information either originated or cited from published or unpublished works of other author have been acknowledged in the text of this thesis and listed in the reference. I also, hereby, give my copyright to Bogor Agricultural University.
Ibaraki, September 2013
SUMMARY
NICHO NURDEBYANDARU. Endophytic Fungi of Wild Zingiberaceae from Mount Halimun Salak National Park. Under supervision of GAYUH RAHAYU, IMAN HIDAYAT and KAZUHIKO NARISAWA.
The genus Colletotrichum, Diaporthe and Guignardia can live as plant pathogens on a wide variety of tropical woody, herbaceous and graminicoulos plants as well as fungal endophytes of many healthy plants. These two life styles are apparently caused by their biotrophic life strategies as such that they live as symptomless endophytes of living plant tissues. The species in Colletotrichum, Diaporthe and Guignardia have been reported to cause serious diseases in various economic plants in Indonesia. The exploration of these fungal endophytes was mainly done on economic plants. Those from non-cultivated plants in natural habitats are much less studied, with most studies being of endophytic strains.No reports are found on fungal endophyte from wild Zingiberaceae plant, therefore it is necessary to conduct research on the endophytic fungi on Zingiberaceae in order to provide information related fungal endophyte of wild Zingiberaceae in Indonesia.
All fungal species recorded from Indonesia were mostly identified based on morphological characters. In this research, isolates of the fungal endophyte was collected by conventional isolation method. The isolates were then identified molecularly based on their ITS1F-ITS4 sequence. These sequences were aligned with MAFFT online. Phylogenetic analyses with all reference strains were performed using MEGA5 to produce the phylogram. The position and bootstrap value for branch support became the criteria of naming the isolates.
Eleven isolates of fungal endophytes were obtained from different part of four species of Zingiberaceae at Mount Halimun Salak National Park (MHSNP) namely Alpinia malaccensis, Ammomum gracile, Etlingera punicea and Zingiber odoriferum. These consisted of nine isolates of Colletotrichum spp., one each isolate of Diaporthe infecunda and Guignardia mangiferae.Three of Colletotrichum isolates were belong to gloeosporioides clade, and six others were separate and made its own clade. The Colletotrichum species distributed widely on those four plant species at different plant part, one isolate of Colletotrichum kahawae subsp. kahawae from root of Etlingera punicea, one isolate of Colletotrichum aff. siamense from root of Alpinia malaccensis, and seven isolates of unidentified Colletotrichum species either from root of Alpinia malaccensis, pseudostem and leaf of Ammomum gracile, and from root of Zingiber odoriferum. One isolate of Diaporthe infecunda was from pseudostem of Alpinia malaccensis and one isolate Guignardia mangiferae was from pseudostem of Zingiber odoriferum. All of this finding is new records on fungal endophytes from wild species of Zingiberaceae and may contribute to the management of domestication of these plants.
v
RINGKASAN
NICHO NURDEBYANDARU. Fungi Endofit dari Zingiberaceae liar asal Taman Nasional Gunung Halimun Salak. Dibimbing oleh GAYUH RAHAYU, IMAN HIDAYAT, dan KAZUHIKO NARISAWA.
Genus Colletotrichum, Diaporthe dan Guignardia dapat hidup sebagai patogen pada berbagai jenis tumbuhan tropis berkayu, herba, rerumputan atau sebagai endofit pada tanaman yang sehat. Colletotrichum, Diaporthe dan Guignardia telah dilaporkan menyebabkan penyakit pada berbagai tanaman bernilai ekonomi tinggi di Indonesia. Eksplorasi fungi endofit ini lebih banyak dilakukan pada tanaman budidaya yang memiliki nilai ekonomi tinggi, sedangkan studi pada tanaman liar sangat jarang dilakukan padahal fungi endofit dilaporkan banyak diisolasi tanaman liar pada habitat aslinya. Beberapa spesies liar Zingiberaceae telah ditemukan pada daerah sekitar Taman Nasional Gunung Halimun Salak (MHSNP). Sebagian dari tanaman tersebut didomestikasi oleh penduduk sekitar karena diduga memiliki kandungan yang bermanfaat. Studi mengenai fungi endofit dari Zingiberaceae liar tersebut belum pernah dilaporkan, oleh karena itu diperlukan penelitian tentang endofit dari Zingiberaceae liar untuk memberikan informasi mengenai keragaman fungi endofit di Indonesia.
Jenis endofit yang telah dilaporkan dari Indonesia umumnya diidentifikasi hanya berdasarkan karakter morfologinya. Pada penelitian ini, fungi diidentifikasi dengan sekuen ITS. Sampel diambil dari bagian tanaman Zingiberaceae yang sehat tanpa penyakit untuk disterilisasi permukaan. Sampel kemudian ditanam di media MEA dan hifa yang tumbuh dari sampel kemudian diisolasi dan dimurnikan pada media yang sama. Isolat murni kemudian diidentifikasi bagian ITS rDNA secara molekular dengan pasangan primer ITS1F dan ITS4. Sekuen ITS disejajarkan secara online dengan program MAFFT. Analisis filogenetik dilakukan dengan MEGA5 dengan membandingkan posisi sekuen isolat dengan sekuen referensi pada filogram.
Sebelas isolat fungi endofit diisolasi dari bagian berbeda dari 4 jenis Zingiberaceae di MHSNP, yaitu Alpinia malaccensis, Ammomum gracile, Etlingera punicea dan Zingiber odoriferum. Sembilan diantaranya yaitu Colletotrichum spp, dan masing-masing satu fungi Diaporthe infecunda dan Guignardia mangiferae. Tiga isolat Colletotrichum tergolong ke dalam clade gloeosporioides, dan sisanya membentuk clade tersendiri. Spesies Colletotrichum tersebut terdistribusi pada empat spesies tanaman pada bagian tanaman yang berbeda. Isolat-isolat tersebut yaitu Colletotrichum kahawae subsp. kahawae dari akar Etlingera punicea, Colletotrichum aff. siamense dari akar Alpinia malaccensis, dan tujuh isolat Colletotrichum lainnya belum teridentifikasi yaitu dari akar Alpinia malaccensis, dari batang dan daun Ammomum gracile, dan dari akar Zingiber odoriferum. Isolat Diaporthe infecunda diisolasi dari batang Alpinia malaccensis dan isolat Guignardia mangiferae diisolasi dari batang Zingiber odoriferum Hasil tersebut diatas merupakan informasi baru mengenai keberadaan fungi endofit dari Zingiberaceae liar di Indonesia yang dapat memberikan kontribusi terhadap manajemen domestikasi tanaman Zingiberaceae tersebut.
© Hak Cipta Milik IPB, Tahun 2013 Hak Cipta Dilindungi Undang-Undang
Dilarang mengutip sebagian atau seluruh karya tulis ini tanpa mencantumkan atau menyebutkan sumbernya. Pengutipan hanya untuk kepentingan pendidikan, penelitian, penulisan karya ilmiah, penyusunan laporan, penulisan kritik, atau tinjauan suatu masalah; dan pengutipan tersebut tidak merugikan kepentingan IPB.
Dilarang mengumumkan dan memperbanyak sebagian atau seluruh karya tulis ini dalam bentuk apa pun tanpa izin IPB.
All Right Reserved IPB Copyright © 2013
Prohibited from quoting any part or all of this paper without including or citing the source. Citations only for educational purposes, research, scientific papers, reports, or review and does not prejudice IPB.
vii
ENDOPHYTIC FUNGI
OF WILD ZINGIBERACEAE FROM
MOUNT HALIMUN SALAK NATIONAL PARK
NICHO NURDEBYANDARU
This thesis is submitted
as a partial fulfillment of the requirement for the
Degree of Master of Science
in Microbiology
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
2013
Research Title : Endophytic Fungi of Wild Zingiberaceae from
Mount Halimun Salak National Park
Name : Nicho Nurdebyandaru
Student Identification Number : G351110211
Study Program : Microbiology
Advisory Board,
Acknowledged by,
Date of examination:
Date of submission:
Dr. Ir. Gayuh Rahayu Supervisor
Dr. Iman Hidayat Co-Supervisor
Dean of Graduate School
Dr Ir Dahrul Syah, MScAgr Coordinator of Microbiology
Study Program
Prof. Dr. Ir. Anja Meryandini
x
PREFACE
I praise Allah for all mercy and gifts so I can complete this research entitled Endophytic Fungi of Wild Zingiberaceae from Mount Halimun Salak National Park. Tributes are addressed to those who contribute to crystallize this work until this thesis was finally accomplished. This research will not success without many parties and individuals helped, therefore I would like to acknowledge Dr. Ir. Gayuh Rahayu, Dr. Iman Hidayat and Prof. Kazuhiko Narisawa as my supervisors for their guidance, time, energy, motivation, advice and valuable support also boundless patience during my study in Indonesia and Japan. My sincere thanks to Dr. Kartini Kramadibrata as external examiner commission and Prof. Dr. Ir. Anja Meryandini as coordinator of Microbiology study program for the advice, input, critic and suggestion until this thesis become better.
I would like to thank to Sepriyadi Rihi S.Si., Israwati Harahap M.Si, and all staff of Mycology Laboratory LIPI-Cibinong who helped so much at this research. Thanks to Mycology IPB lab member: Eris Septiana, Ivan Permana Putra, Tatik Hartanti, Kemala S. Nagur, for togetherness, discussion and motivation during first year at Indonesia, also Oktan Dwi Nurhayat S.Si., Erwin S.Pd., for their time and support at Mycology laboratory of IPB. Rola Sameer Mahmoud, Yuki Komatsuzaki, Erika Usui, Saki Shingaki and Surono for their support, help and discussion during this research in Japan. I also thanks to Prof. Nobuo Sakagami for all the administrative assistance.
I want to thank my father, Dr. Ir. Rubiyo M.Si., my mother Ir. Endang Mufrihati, my brother Briliandaru MP S.T., Dhimas Upadyandaru, Dhaksa Buwana, my sister Azzalia and Adelia, and my fiancé Angelia Rezty for their patience, love and pray.
I hope this paper benefiting others.
Ibaraki, September 2013
CONTENTS
SUMMARY iv
PREFACE x
CONTENT xi
LIST OF TABLES xii
LIST OF FIGURES xii
LIST OF APPENDIXES xii
1 INTRODUCTION 1
Background 2
Objectives 2
Hypothesis 2
Uses 2
2 MATERIAL AND METHOD Time and Place 3
Plant Material 3
Isolation of Endophytic Fungi 3
DNA Extraction, Amplification and Sequencing of Fungal ITS 4
Phylogenetic Analysis 4
Morphology Observation 5
3 RESULT AND DISCUSSION Endophytic Species of Colletotrichum 5
Endophytic Species of Diaporthe 14
Endophytic Species of Guignardia 19
4 CONCLUSION 20
REFERENCES 21
APPENDIX 23
xii
LIST OF TABLES
1 List of reference Colletotrichum strains and out group used in this study 6
2 The shape and size of Colletotrichum appressoria (Weir et al. 2012) 14
3 List of reference Diaporthe strain used in this study 15
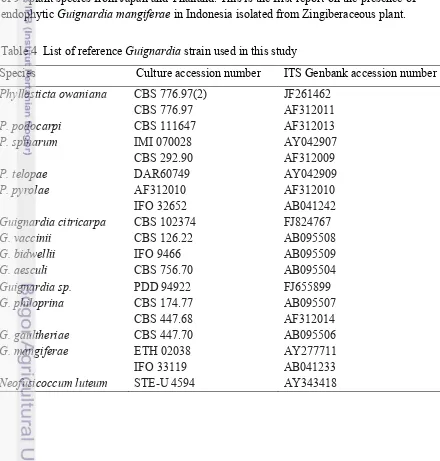
4 List of reference Guignardia strain used in this study 19
LIST OF FIGURES
1 Habitus of Alpinia malaccensis (A), Ammomum gracile (B), Etlingera punicea (C) and Zingiber odoriferum (D) 3 2 Phylogramme of Colletotrichum derived from an ML analyses of ITS1F-ITS4(+500bp) sequence using Kimura-2 model, with gamma distribution (G) and
evolutionary invariable (I), bootstrap 1000x 11
3 Subtrees of Colletotrichum gloeosporioides complex with C. musae subclade
(A) and C. kahawae subclade (B) 13
4 Hyphal appressoria (red arrow) of the isolate A) 133A1, B) 511A2, C)
611A4A1 and D) 6Fr2C1 13
5 Phylogramme of Diaporthe derived from an ML analyses of ITS1F-ITS4 (+500bp) sequence using Tamura-Nei model, with gamma distribution (G) and
evolutionary invariable (I), bootstrap 1000x 17
6 Phylogramme of Guignardia derived from ML analyses of ITS1F-ITS4 (+500bp) sequence using kimura-2 model, with gamma distribution (G) and
bootstrap 1000x 20
LIST OF APPENDIXES
1 Maximum Likelihood (ML) of 24 different nucleotide substitution models for
Colletotrichum species 23
2 Maximum Likelihood (ML) of 24 different nucleotide substitution models for
Diaporthe species 24
3 Maximum Likelihood (ML) of 24 different nucleotide substitution models for
1 INTRODUCTION
Background
Endophytic fungi have been defined as fungi that inhabit host plant tissue in their life cycle without causing visible symptoms of disease (Schulz & Boyle 2005). This group of fungi can be found in every part of the plant and also in various kinds of plants (Rodriguez et al. 2009). Every plant examined to date harbors at least one species of endophytic fungus. Many plants, especially woody plants, may contain hundreds or thousands of fungal species. Arnold et al. (2000) for example, isolated more than a thousand fungal strains that consist of 418 endophytic morphospecies from leaves of Heisteria concinna (Olaceae) and Ouratea lucens (Ochnaceae) at tropical rain forest of Panama. Huang et al. (2008) reported that they were able to get 1,160 fungal isolates representing 31 taxonomic groups, including 73 morphospecies from 29 Chinese traditional medicinal plants. Taxa from Alternaria, Colletotrichum, Phoma, Phomopsis and Xylariales are the dominant fungal endophytes. In Indonesia, Ilyas et al. (2009) isolated 53 isolates of endophytic fungi from Uncaria gambier Roxb. (Rubiaceae), a spice plant, in West Sumatra. These endophytic fungi belong to either Coelomycetes such as Phomopsis, Pestalotiopsis, Phoma or Hypomycetes, such as Aspergillus, Cladosporium, Fusarium and Penicillium.
Zingiberaceae is known as one of medicinal and spices plants that expectedly to harbor high number of endophytic fungi. This family is distributed mainly in tropical Asia (South and Southeast), America (Wu & Larsen 2000) and also Africa (Dhetchuvi et al. 2011) with its greatest diversity in Southeast Asia. However, information regarding endophytic fungal diversity isolated from zingiberaceous plants is scarce. Several reports have been recorded from Southeast Asia, for example, 36 species of fungal endophytes has been reported to associate with wild ginger, Ammomum siamese (Zingiberaceae) from Thailand (Bussaban et al. 2001). Of those endophytes, some coelomyctes such as Colletotrichum gloeosporoides and Phomopsis spp. were regarded as the dominant fungi. They found two new Ascomycetes species, Gaeumannomyces amomi and Leiosphaerella amomi, and three new species of Pyricularia and found other fungi such as Aspergillus, Cladosporium, Diaporthe, Fusarium and Penicillium. They also noted that Fusarium spp. occured from different plant parts such as pseudostem and rhizome of Ammomum siamense (Bussaban et al. 2001).
2
Although Indonesia is rich of Zingiberaceae biodiversity, thorough information on its pathogenic and endophytic diversity and fungal community profile is still lacking. Noverita et al. (2009) was the only report of fungal endophytes on cultivated Zingiberaceae from Indonesia. They only studied on Zingiber ottensii (Bangle ghost), and were able to isolate 10 endophytic fungi from leaf and rhizome without further identification. Above all, Indonesia has about 650 species, of which ten species of Zingiberaceae are found in Mount Halimun Salak National Park (MHSNP) (Priyadi et al. 2010). Some of those species are considered as wild plants and under domestication process. The information on endophyte of these wild zingiberaceous plant is lacking.
The information on diversity of endophytic fungi on Zingiberacae is important as bases for the Zingiberaceae domestication strategy. There are possibilities of changes from biotrophic to necrotrophic life style in Colletotrichum graminicola (Thon et al. 2002). Elucidation of fungal endophytes assemblages on various host plants using conventional method through isolation (Guo et al. 1998) has been reported successful. Therefore, this method will be used to explore the endophyte associated with wild Zingiberaceae from MHSNP.
Research Objective
This study is aimed to study endophytic fungi diversity some Zingiberaceae from MHSNP.
Hypothesis
1. Wild Zingiberaceae hosted many endophytic fungi. 2. Some of endophytic fungi belong to coelomycetes.
Uses
1. First report of endophytic fungi from wild species of Zingiberaceae in Indonesia. 2. Contribute to the knowledge of fungal biodiversity in Indonesia
2
METHODS
Time and Place
The research had been conducted from March 2012 - August 2013 at the laboratory of Mycology, Division of Mycology, Department of Biology, Bogor Agricultural University and for sequencing at Gene Research Center (GRC), Ibaraki University.
Plant material
Four species of healthy wild Zingiberaceae were collected from Gunung Halimun Reserve Area (1086-1134 m asl). These plants namely Alpinia malaccensis were found at S: 06° 44' 27.7", E: 106° 31' 46.8", Ammomum gracile (S: 06° 44' 27.5", E: 106° 31' 46.1"), Etlingera punicea (S: 06° 44' 24.4", E: 106° 31' 49.3") and Zingiber odoriferum (S: 06° 44' 27.7", E: 106° 31' 46.8") (Figure 1). All plants were identified morphologically.
Figure 1 Habitus of Alpinia malaccensis (A), Ammomum gracile (B), Etlingera punicea (C) and Zingiber odoriferum (D).
Isolation of Endophytic Fungi
A total of 10 samples of plant parts (leaf, stem, flower, fruit and root) from each species were first washed with running tap water. The plant parts were cut into 5 x 5 cm pieces to be surface sterilized with 0.1% clorox for 2 minutes followed by 70% alcohol for 1 minute before rinsing with distilled water for 3 times. These pieces were then dried up by placing over the sterile tissue paper in sterile Petri dishes for at least 6 hours. Five pieces of one plant part were placed onto half strength MEA agar and kept for a month in room temperature. Whitish to dark colonies were selected to be purified. Pure cultures were then maintained on Potato Dextrose Agar (Oxoid) slants.
4
DNA Extraction, Amplification and Sequencing of Fungal ITS
Five days old colony grown on Potato Dextrose Broth was used as a DNA source. Isolates were selected for molecular identification. Fungal genomic DNA was prepared with the NucleonTMPhytoPureTM (GE Healthcare, UK) extraction kit following the manufacturer’s protocol. Amplification of rDNA ITS region on samples was carried out by PCR machine using universal primer pair ITS1F (CTTGGTCATTTAGAGG) (Gardes & Bruns 1993) and ITS4 (CCTCCGCTTATTG) (White et al. 1990). For each sample, amplification was performed in a 50 µl reaction volume, which contained 5µl PCR buffer, 4 µl dNTPS, 2.5 µl each primer, 0.15 µl Taq DNA polimerase (Takara), 0.5 µl DNA template and were added with miliQ water until 50 µl volume. Thermal cycle consisted of 5 minutes initial denaturation at 94˚C, followed by 35 cycles of 35 seconds denaturation at 94˚C, 55 seconds primer annealing at 55˚C, 2 minutes extension at 72˚C, and a final 10 min extension at 72˚C. PCR products were kept at 4˚C. Five µl of PCR products from each PCR reaction were examined by electrophoresis in a 1% agarose gel in 1×TAE buffer at 100 V for 20 minutes and visualized under UV light after being stained with GelRed (Wako Pure Chemical, Japan).
PCR products were purified by adding 43.5 µl of PCR purification solution (12 µl 3M CH3COONa pH 4.8, 30 µl 40% PEG, and 1.5 µl 200 mM MgCl2) into each PCR product tube and these solutions were vortexed and incubated at 4˚C overnight. After incubation time solution were centrifuged at 15000 rpm at 4˚C for 15 minutes, and supernatant were removed. About 180 µl 80% ethanol were added and solution were centrifuged at 15000 rpm at 4˚C for 15 minutes also supernatant were removed. Absolute ethanol were then added and centrifuged with the same condition as mentioned above. Sequencing PCR was done by adding 0.5 µl DNA template onto 9.5 µl solution system (1.5 µl Buffer, 0.5 µl BigDye, 0.32 µl primer ITS1F, and miliQ water until volume reached 9.5 µl) in 0.2 ml PCR tube. Sequencing PCR cycle were set as 2 minutes for initial denaturation at 96˚C, followed by 25 cycles of 30 second denaturation at 96˚C, 15 second primer annealing at 50˚C, and 3 minute extension at 60˚C.
Purification of sequencing product were done by adding purification solution (12 µl 3M CH3COONa pH 4.8, 30 µl 40% PEG, and 1.5 µl 200 mM MgCl2) three times of volume of sequencing product. The solutions were centrifuged at 15000 rpm at 4˚C for 20 minutes and supernatant were removed. A hundred fifty µl of ethanol 70% were added and were centrifuged as the same as above. Pellet were dried and covered by tissue paper at room temperature. About 20 µl HIDI were added into each pellet and were vortexed at 2000 rpm for 2 minutes, then solution were pipetted into the 96-cell-plate and were analyzed into sequencing machine ABI 3130 Genetic Analyzer (Applied Biosystem).
Phylogenetic Analysis
internal branches was estimated by bootstrap analysis based on 1,000 replications. Monilichaetes infuscans CBS 869.96, Diaporthella corylina CBS 121124 and Neofusicoccum luteum STE-U 4594 were used as an out-group, respectively for Colletotrichum, Diaporthe and Guignardia.
Morphology Observation
A single colony from culture stock was grown at MEA and PDA in a 9 mm Petridishes and incubated at room temperature until the sporulation was observed. All reproductive structures if present were noted.
3 RESULT AND DISCUSSION
Totally eleven white to dark isolates were collected from Zingiberaceae from MHSNP. These isolates showed different morphological features. However, it is not possible to determine further to species level since all this isolates were unsporulated. In the beginning of the isolation process some of these isolates showed orange pustules that indicate the acervuli of the Colletotrichum. Some others showed distinctive fruit body without spores in it. When the ITS sequence from these isolates were aligned to the Genbank data, most of the isolates showed highly similarity with Colletotrichum gleosporioides, one isolates each with either Colletotrichum boninense or Colletotrichum crassipes. The two other isolates, one is similar to a different species of Diaporthe (D. longicola and D. helianthi (100%) or D. chimonanti and D. infecunda (99%) and another isolate with Guignardia vaccini (100%). These identification results were further optimized and checked for gaining more reliable result. Analyses the identities of the isolates to species level were made.
Endophytic Species of Colletotrichum/Glomerella (Teleomorph/Anamorph)
Of those isolates with white to dark colonies, nine isolates are apparently Colletotricum. These composed of four isolates from root, pseudostem and fruit of Alpinia malaccensis, two isolates from pseudostem and leaf Ammomum gracile, one isolate from root of Etlingera punicea, and two isolates from root of Zingiber odoriferum. These isolates showed orange pustules during first step of isolation. However, the capability of forming conidia and acervuli apparently lost during purification.
6
in the Genbank might be incorrectly identified to species level. In particular, Hyde et al. (2010) stated that sequence lodged in Genbank, as C. gloeosporioides were doubtful and belonged to many different species. Further, those ITS sequences of C. gloeosporioides and Glomerella cingulata (teleomorph of C. gloeosporioides) were found throughout the C. gloeosporioides species complex and outside the C. gloeosporioides complex. Those outside C. gloeosporioides applied to more than 100 of the about 750 ITS sequences of C. gloeosporioides in GenBank. Therefore, identification of these isolates used phylogenetic analyses.
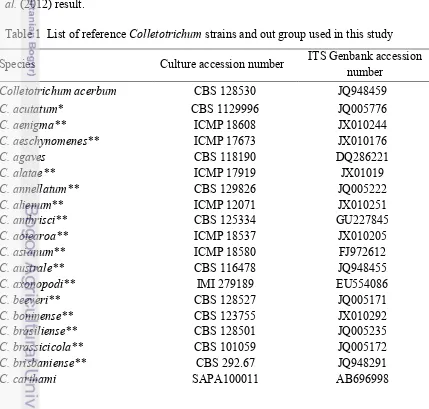
The alignment in the phylogenetic analyses of the isolates collected, included sequences from all species of Colletototrichum were reported by Cannon et al. (2012) of which all are either holotypes or epitypes (Table 1). The data was analyzed by maximum likelihood (ML), which performed the clades that meet with that of Cannon et al. (2012). All nine clades in the Colletotrichum complex were shown in the phylogram eventhough they are supported by low bootstrap value, except for C. truncatum and C. orbiculare clade (Figure 2). The branches of these clades were supported by 87 and 99% bootstrap values, respectively. However, Cannon et al. (2012) stated that C. gloeosporioides complex is one amongst nine well-established major clades species in the genus Colletotrichum. In this analysis, C. gloeosporioides complex separated from the other species complex or clades similar with Cannon et al. (2012) result.
Table 1 List of reference Colletotrichum strains and out group used in this study
Species Culture accession number ITS Genbank accession number
Colletotrichum acerbum CBS 128530 JQ948459
C. acutatum* CBS 1129996 JQ005776
C. aenigma** ICMP 18608 JX010244
C. aeschynomenes** ICMP 17673 JX010176
C. agaves CBS 118190 DQ286221
C. alatae** ICMP 17919 JX01019
C. annellatum** CBS 129826 JQ005222
C. alienum** ICMP 12071 JX010251
C. anthrisci** CBS 125334 GU227845
C. aotearoa** ICMP 18537 JX010205
C. asianum** ICMP 18580 FJ972612
C. australe** CBS 116478 JQ948455
C. axonopodi** IMI 279189 EU554086
C. beeveri** CBS 128527 JQ005171
C. boninense** CBS 123755 JX010292
C. brasiliense** CBS 128501 JQ005235
C. brassicicola** CBS 101059 JQ005172
C. brisbaniense** CBS 292.67 JQ948291
C. cereale CBS 129663 DQ126177
C. chlorophyti** IMI 103806 GU227894
C. chrysanthemi SAPA 100010 AB696999
C. circinans CBS 221.81 GU227855
C. clidemiae** ICMP18658 JX010265
C. cliviae CBS 125375 GQ485607
C. coccodes CBS 369.75 HM171679
C. colombiense** CBS 129818 JQ005174
C. constrictum CBS 128504 JQ005238
C. cosmi CBS 853.73 JQ948274
C. costaricense** CBS 330.75 JQ948180
C. curcumae IMI 288937 GU227893
C. cuscutae** IMI 304802 JQ948195
C. cymbidiicola** IMI 347923 JQ005166
C. cordylinicola** ICMP18579 JX010226
C. dacrycarpi** CBS 130241 JQ005236
C. dematium* CBS 125.25 GU227819
C. destructivum CBS 149.34 AJ301942
C. dracaenophilum** CBS 118199 DQ286209
C. echinochloae MAFF 511473 AB439811
C. eleusines MAFF 511155 EU554131
C. eremochloae** CBS 129661 JQ478447
C. falcatum*** CBS 147945 JQ005772
C. fioriniae CBS 128517 JQ948292
C. fructi CBS 346.37 FJ972603
C. fructicola ICMP 172921 JX010165
C. fuscum CBS 130.57 JQ005762
C. gloeosporioides* ICMP 17821 JX010152
C. godetiae** CBS 133.44 JQ948402
C. graminicola* CBS 130836 DQ003110
C. guajavae** IMI 350839 JQ948270
C. hanaui** MAFF 305404 JX519217
C. hemerocallidis** CDLG5 JQ400005
C. higginsianum IMI 349063 JQ005760
C. hipperastri CBS 241.78 JQ005231
C. horii*** ICMP 10492 GQ329690
C. indonesiense** CBS 127551 JQ948288
C. jacksonii MAFF 305460 EU554108
C. jasminigenum** MFLUCC 10 0273 HM1311513
C. johnstonii CBS 128532 JQ948444
C. karstii CBS 132134 HM585409
C. kahawae subsp. kahawae** ICMP 17816 JX010231
8
C. kinghornii** CBS 198.35 JQ948454
C. laticiphilum** CBS 112989 JQ948289
C. lilii CBS 109214 GU227810
C. limetticola* CBS 114.14 JQ948193
C. lindemuthianum CBS 144.31 JQ005779
C. lineola* CBS 125337 GU227829
C. linicola CBS 172.51 JQ005765
C. liriopes** CBS 119444 GU227804
C. lupine*** CBS 109225 DQ286119
C. melonis** CBS 159.84 JQ948194
C. miscanthi** MAFF 510857 EU554121
C. musae* ICMP 19119 JX010146
C. navitas** CBS 125086 GQ919067
C. nicholsonii** MAFF 511115 EU554126
C. novae-zelandiae** CBS 128505 JQ005228
C. nupharicola** ICMP 18187 JX010187
C. nymphaeae* CBS 515.78 JQ948197
C. oncidii** CBS 129828 JQ005169
C. orchidophilum** CBS 632.80 JQ948151
C. orbiculare CBS 514.97 JQ005778
C. parsonsiae** CBS 128525 JQ005233
C. paspali** MAFF 305403 EU554100
C. paxtonii** IMI 165753 JQ948285
C. petchii* CBS 378.94 JQ005223
C. phaseolorum# CBS 157.36 GU227896
C. phormii* CBS 118194 JQ948446
C. phyllanthi** CBS 175.67 JQ005221
C. pseudoacutatum** CBS 436.77 JQ948480
C. psidii# ICMP 19120 JX010219
C. pyricola** CBS 128531 JQ948445
C. quenslandicum* ICMP 1778 JX010276
C. rhombiforme** CBS 129953 JQ948457
C. rusci** CBS 119206 GU227818
C. salicis* CBS 607.94 JQ948460
C. salsolae** ICMP 19051 JX010242
C. sansevieriae** MAFF 239721 AB212991
C. scovillei** CBS 126529 JQ948267
C. siamense** ICMP 18578 JX010171
C. simmondsii** CBS 122122 JQ948276
C. sloanei** IMI 364297 JQ948287
C. spaethianum* CBS 167.49 GU227807
C. spinaciae CBS 128.57 GU227847
C. tabacum CBS 161.53 JQ005763
C. theobromicola*** ICMP 18649 JX010294
C. ti** ICMP 4832 JX010269
C. tofieldiae CBS 495.85 GU227801
C. torulosum** CBS 128544 JQ005164
C. trichellum CBS 217.64 GU227812
C. tropicale** ICMP 18653 JX010264
C. truncatum* CBS 151.35 GU227862
C. verruculosum** IMI 45525 GU227806
C. walleri** CBS 125472 JQ948275
C. xanthorrhoeae BRIP 45094 GU048667
C. yunnanense** CBS 132135 EF369490
Monilochaetes infuscans CBS 869.96 GU180626.1
* : epitype, ** : ex-holotype, *** : ex-neotype, # : authentic culture
The phylogram of the ITS region showed that the nine isolates Colletotrichum distributed into 2 different clades i.e. C. gloeosporioides and a presumably new clades (Figure 2).Identification further within clades is problematic. Weir et al. (2012) also stated that ITS can only be used to separate clades.
The isolates in C. gloeosporioides clades were belong to isolate 611A4, 133A1 and 6Fr2C1. Two verification approaches were taken to confirm the position of these isolates in the C. gloeosporioides complex. The first verification used unique string of C. gloeosporioides i.e 5’-GGGCGGGT-3’. According to Weir et al. (2012) about 139-142 bases after the ITS1F primer binding site, all of the isolates accepted in the C. gloeosporioides complex share the string 5’-GGGCGGGT-3’. They also found this string appeared to be specific to isolates of the C. gloeosporioides complex on the bases of their comparison with GenBank data. In this study only two isolates (133A1 and 611A4) have such kind of string proofing that the isolates are members of gloeosporioides complex. In contrast, isolates 6Fr2C1 did not have such a string.
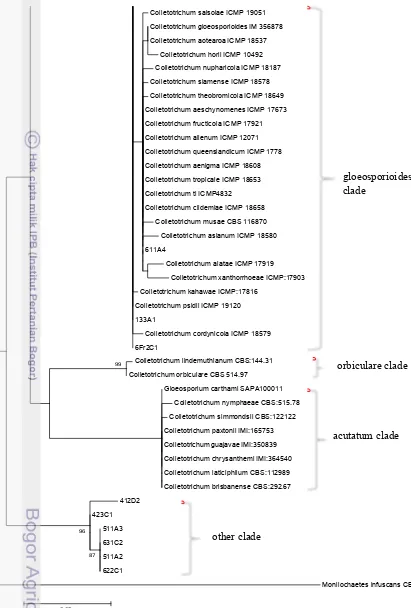
The isolates that are in C. gloeosporioides clades belong to two subclades, i.e. C. musae (611A4) and C. kahawae (133A1 and 6Fr2C1) subclades (Figure 3). The position of isolate in the C. musae clades cannot be determined yet since the branch has low supported bootstrap value (39%). Therefore isolates 133A1 is tentatively identified as Colletotrichum aff. siamense. A low bootstrap value (37%) also supported the branch of kahawae subclade. Therefore, those in kahawae should be determined by comparison using the available morphological data.
10
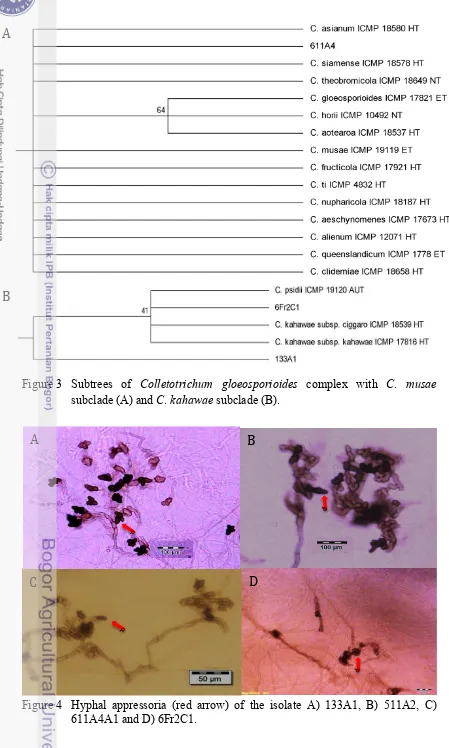
complex, Weir et al. (2012) only gave the illustration and description of the hyphal appressoria of eight member of C. gloeosporioides complex (Table 2). Comparison to the illustration of appressoria confirmed that isolate 133A1 is C. kahawae subsp. kahawae. The hyphal appressoria of 6Fr2C1 did not meet with any illustration in kahawae subclades, thus identification to species level cannot be made. Hyphal appressoria of 611A4 did not meet with any illustration available for species in musae subclade as well and thus cannot confirm the position as Colletotrichum aff. siamense. The appressoria of isolate 133A1 are mostly globose with slightly lobed, whilst that of 511A2 and 6Fr2C1 are mostly lobed and lumped (Figure 4). The available information on the appressoria (Tabel 2) is poor, thus it cannot be used as a key factor to differentiate the species. Therefore, confirmation using other markers is needed.
The other six isolates (412D2, 423C1, 511A2, 511A3, 622C1 and 631C2) are grouped forming a strong clade with 96% bootstrap value. There is a possibility that this clade is a new clade and new species. All the isolates also showed hyphal appressoria. However, this could only confirm the isolates until genus level. Confirmation into species level using other markers is needed.
This study reveals new result of the presence of endophytic Colletotrichum from Zingiberaceae from MHSNP. Colletotrichum kahawae subsp. kahawae is a new record in Indonesia. In Africa, C. kahawae subsp. kahawae is an important pathogen of coffee berry (Weir et al. 2012) and has never been reported as an endophyte. This evidence of endophytic Colletotrichum species confirmed the report of Bussaban et al. (2001) who found the endophytic C. gloeosporioides from wild species of Zingiberaceae (Ammomum siamense) and Photita et al. (2005) who found that species in Alpina malaccensis in Thailand.
This study found that one individual plant might harbor more than one species of Colletotrichum. Alpinia malaccensis hosted Colletotrichum cf. siamense on root and two of unidentified diferrent lineage of Colletotrichum in the fruit and pseudostem. Ammomum gracile hosted presumably the same species on their pseudostem and leaf. Freeman and Katan (1997) reported that Colletotrichum has been known to infect all plant part above the ground and also root.
12
Figure 2 Phylogramme of Colletotrichum derived from an ML analyses of ITS1F-ITS4 (+500bp) sequence using Kimura-2 model, with gamma distribution (G) and evolutionary invariable (I), bootstrap 1000x.
Figure 3 Subtrees of Colletotrichum gloeosporioides complex with C. musae subclade (A) and C. kahawae subclade (B).
Figure 4 Hyphal appressoria (red arrow) of the isolate A) 133A1, B) 511A2, C) 611A4A1 and D) 6Fr2C1.
A
B
C
D
A
14
Table 2 The shape and size of Colletotrichum appressoria (Weir et al. 2012)
Species Shape Size
Colletotrichum
aenigma Subglobose or with a few broad lobes 6-10 µm C. aeschynomenes Mostly elliptic to subfusoid, deeply
lobed. -
C. alatae
Mostly simple, elliptic to fusoid in shape, sometime developing broad, irregular cylindric, a few with broad, irregular lobes
C. atoteroa
Variable in shape, simple to broadly lobed, sometimes in groups, sometimes intercalary,
7–17 × 4–9.5 µm
C. clidemiae
Variable in shape, some simple, subglobose, but often with a small number of broad, irregular lobes C. crassipes Appressoria deeply lobed. C. kahawae subsp.
ciggaro
Typically cylindric to fusoid in shape, deeply lobed
C. kahawae subsp.
kahawae *
C. queenslandicum Globose to short-cylindric, rarely lobed 6–12 µm in φ *only illustration available
Endophytic Species of Diaporthe
In this study the identification of Diaporthe is only used ITS region, eventhough according to Gomes et al. (2013), the locus could distinguish only 75 of the 95 species (79 % success). They suggested that identification should be based on multigene, a combination of ITS, TEF1, TUB, HIS and CAL region. All Diaporthe
species recognized by Gomes et al. (2013) were used as reference strains (Table 3) with Diaporthella corylina as outgroup. Phylogenetic study of the isolate originated from pseudostem of Alpinia malaccensis indicated that the isolate(6Fr2C3) belongs to Diaporthe infecunda with 86% bootstrap value (Figure 5). Gomes et al. (2013) also stated that D. infecunda has ITS positions 108 (T), 279 (C), 292 (G), 359 (C) and 360 (G). Yet, confirmation using this character has not been done. The isolate being studied has no reproductive structure and this evidence is similar to that of Gomes et al. (2013), which was also sterile.
The presence of D. infecunda on Zingiberaceae in Indonesia is the first report. The specimens of D. infecunda studied by Gomes et al. (2013) were also endophyte
petiole of Maytenus ilicifolia (Celastraceae), and from leaves of Schinus terebinthifolius (Anacardiaceae). Endophytic Diaporthe sp. has also found on
Cinchona ledgeriana in Indonesia (Maehara et al. 2012). Other than endophytic
species, pathogenic species of Diaporthe, Diaporthe phaseolorum (Gomes et al.
2013) and Diaporthe citri had been reported to occur on Glycine max and unknown plant, respectivelyin Indonesia.
Table 3 List of reference Diaporthe strain used in this study
Species Culture accession
number
ITS Genbank accession number Diaporthe acaciigena* CBS 129521 KC343005
D. acerina CBS 137.27 KC343006
D. alleghaniensis* CBS 495.72 KC343007
D. alnea CBS 146.46 KC343008
D. ambigua** CBS 114015 KC343010
D. ampelina STE-U 2673 KC343016
D. amygdali CBS 111811 KC343019
D. angelicae** CBS 111592 KC343027
D. arctii CBS 136.25 KC343031
D. arecae*** CBS 161.64 KC343032
D. arengae* CBS 114979 KC343034
D. aspalathi* CBS 117169 KC343036
D. australafricana* CBS 111886 KC343038
D. batatas CBS 122.21 KC343040
D. beckhausii CBS 138.27 KC343041
D. brasiliensis* CBS 133183 KC343042
D. carpini CBS 114437 KC343044
D. caulivora**** CBS 127268 KC343045
D. celastrina CBS 139.27 KC343047
D. chamaeropis CBS 454.81 KC343048
D. cinerascens CBS 719.96 KC343050
D. citri CBS 230.52 KC343052
D. conorum CBS 186.37 KC343079
D. convolvuli CBS 124654 KC343054
D. crataegi CBS 114435 KC343055
D. crotalariae* CBS 162.33 KC343056
D. cuppatea* CBS 117499 KC343057
D. cynaroidis* CBS 122676 KC343058
D. decedens CBS 109772 KC343059
D. detrusa CBS 109770 KC343061
D. elaeagni CBS 504.72 KC343064
D. endophytica* CBS 133811 KC343065
D. eres CBS 101742 KC343074
D. eugeniae CBS 444.82 KC343098
D. fibrosa CBS 109751 KC343099
D. foeniculacea CBS 111553 KC343101
D. ganjae* CBS 180.91 KC343112
16
D. helianthi* CBS 592.81 KC343115
D. cf. heveae 1 CBS 852.97 KC343116
D. hickoriae* CBS 145.26 KC343118
D. hongkongensis* CBS 115448 KC343119
D. hordei CBS 481.92 KC343604
D. impulsa CBS 114434 KC343121
D. inconspicua* CBS 133813 KC343123
D. infecunda* CBS 133812 KC343126
D. juglandina CBS 121004 KC343134
D. longispora* CBS 194.36 KC343135
D. lusitanicae* CBS 123212 KC343136
D. manihotia CBS 505.76 KC343138
D. mayteni* CBS 133185 KC343139
D. megalospora CBS 143.27 KC343140
D. melonis*** CBS 507.78 KC343142
D. musigena* CBS 129519 KC343143
D. neilliae CBS 144.27 KC343144
D. neoarctii* CBS 109490 KC343145
D. neotheicola* CBS 123209 KC343105
D. nobilis CBS 113470 KC343146
D. nobilis CBS 200.39 KC343151
D. nomurai CBS 157.29 KC343154
D. novem* CBS 127270 KC343156
D. oncostoma CBS 100454 KC343160
D. oxe* CBS 133186 KC343164
D. padi var. padi CBS 114200 KC344137
D. paranensis* CBS 133184 KC343171
D. perjuncta** CBS 109745 KC343172
D. perseae CBS 151.73 KC343173
D. phaseolorum CBS 116019 KC343175
D. pseudomangiferae* CBS 101339 KC343181
D. pseudophoenicicola CBS 176.77 KC343183
D. pustulata CBS 109742 KC343427
D. raonikayaporum* CBS 133182 KC343188
D. rhoina CBS 146.27 KC343189
D. saccarata* CBS 116311 KC343190
D. schini* CBS 133181 KC343191
D. sclerotioides* CBS 296.67 KC343193
D. scobina CBS 251.38 KC343195
D. sojae CBS 100.87 KC343196
D. stictica CBS 370.54 KC343212
D. subordinaria CBS 101711 KC343213
D. tecomae CBS 100547 KC343215
D. terebinthifolii* CBS 133180 KC343216
D. toxica* CBS 534.93 KC343220
D. vaccinii* CBS 160.32 KC343228
D.vexans CBS 127.14 KC343229
D. viticola CBS 100170 KC343230
Diaporthella corylina CBS 121124 KC343004
Diaporthe cynaroidis strain CBS 122676 Diaporthe australafricana strain CBS 111886 Diaporthe australafricana strain CBS 113487
Diaporthe beckhausii strain CBS 138 27 Diaporthe toxica strain CBS 534 93 Diaporthe toxica strain CBS 535 93
Diaporthe padi var padi strain CBS 114200 Diaporthe padi var padi strain CBS 114649
Diaporthe saccarata strain CBS 116311
Diaporthe cf heveae 1 RG 2013 strain CBS 852 97 Diaporthe caulivora strain CBS 127268
Diaporthe caulivora strain CBS 178 55
Diaporthe sp 5 RG 2013 strain CBS 125575 Diaporthe brasiliensis strain CBS 133183 Diaporthe scobina strain CBS 251 38
Diaporthe ampelina strain CBS 111888 Diaporthe ampelina strain STE U2660
Diaporthe impulsa strain CBS 141 27
Diaporthe crataegi strain CBS 114435 Diaporthe nomurai strain CBS 157 29 Diaporthe woolworthii strain CBS 148 27
Diaporthe detrusa strain CBS 109770 Diaporthe detrusa strain CBS 114652 Diaporthe acerina strain CBS 137 27
Diaporthe crotalariae strain CBS 162 33 Diaporthe aspalathi strain CBS 117168
Diaporthe aspalathi strain CBS 117169 Diaporthe woodii strain CBS 558 93
Diaporthe amygdali strain CBS 126679
Diaporthe hickoriae strain CBS 145 26 Diaporthe cinerascens strain CBS 719 96
Diaporthe anacardii strain CBS 720 97 Diaporthe inconspicua strain LGMF922 Diaporthe inconspicua strain CBS 133813
Diaporthe elaeagni strain CBS 504 72 Diaporthe stictica strain CBS 370 54 Phomopsis sp strain RJGD D16
Diaporthe foeniculacea strain CBS 116957 Diaporthe foeniculacea strain CBS 123208 Diaporthe chamaeropis strain CBS 454 81
Diaporthe chamaeropis strain CBS 753 70 Diaporthe celastrina strain CBS 139 27
Diaporthe juglandina strain CBS 121004
Diaporthe alleghaniensis strain CBS 495 72 Diaporthe eres strain CBS 439 82
Diaporthe eres strain CBS 445 62 Diaporthe vaccinii strain CBS 122116 Diaporthe vaccinii strain CBS 160 32
Phomopsis sp strain RJGD D9 Diaporthe gardeniae strain CBS 288 56 Diaporthe cf nobilis RG 2013 strain CBS 113470
Diaporthe cf nobilis RG 2013 strain CBS 116953 Diaporthe alnea strain CBS 146 46
Diaporthe neilliae strain CBS 144 27
Diaporthe rhoina strain CBS 146 27
Diaporthe pseudophoenicicola strain CBS 176 77 Diaporthe pseudophoenicicola strain CBS 462 69
Diaporthe sp 7 RG 2013 strain CBS 458 78 Diaporthe sp 8 RG 2013 strain LGMF925
Diaporthe arecae strain CBS 161 64
Diaporthe cf heveae 2 RG 2013 strain CBS 681 84 Diaporthe hongkongensis strain CBS 115448
Diaporthe arecae strain CBS 535 75 Diaporthe perseae strain CBS 151 73
Diaporthe sp 6 RG 2013 strain CBS 115584
Diaporthe musigena strain CBS 129519 Diaporthe arengae strain CBS 114979
Diaporthe pseudomangiferae strain CBS 101339
Diaporthe eugeniae strain CBS 444 82 Diaporthe sp 3 RG 2013 strain CBS 287 29
18
Figure 5. Phylogramme of Diaporthe derived from an ML analyses of ITS1F-ITS4 (+500bp) sequence using Tamura-Nei model, with gamma distribution (G) and evolutionary invariable (I), bootstrap 1000x.
Diaporthe longispora strain CBS 194 36
Diaporthe strumella var longispora isolate CBS 194 36 Diaporthe sclerotioides strain CBS 296 67 Diaporthe sclerotioides strain CBS 710 76 Diaporthe lusitanicae strain CBS 123212 Diaporthe lusitanicae strain CBS 123213 Diaporthe batatas strain CBS 122 21
Diaporthe sp 2 RG 2013 strain LGMF932 Diaporthe raonikayaporum strain CBS 133182 Diaporthe schini strain CBS 133181
Diaporthe infecunda strain LGMF908 Diaporthe infecunda strain CBS 133812 6Fr2C3
Diaporthe helianthi strain CBS 344 94 Diaporthe helianthi strain CBS 592 81 Diaporthe megalospora strain CBS 143 27
Diaporthe melonis strain CBS 435 87 Diaporthe melonis strain CBS 507 78
Diaporthe sojae strain CBS 100 87 Diaporthe sojae strain CBS 116017
Diaporthe tecomae strain CBS 100547 Diaporthe ganjae strain CBS 180 91 Diaporthe manihotia strain CBS 505 76 Diaporthe sp 1 RG 2013 strain CBS 119639 Diaporthe phaseolorum strain CBS 113425 Phomopsis dauci isolate CBS 315 49 Diaporthe angelicae strain CBS 123215 Diaporthe angelicae strain CBS 111592
Diaporthe subordinaria strain CBS 464 90 Diaporthe arctii strain CBS 136 25 Diaporthe stewartii isolate CBS 193 36
Diaporthe neoarctii strain CBS 109490 Diaporthe hordei strain CBS 481 92 Diaporthe convolvuli strain CBS 124654
Endophytic Species of Guignardia
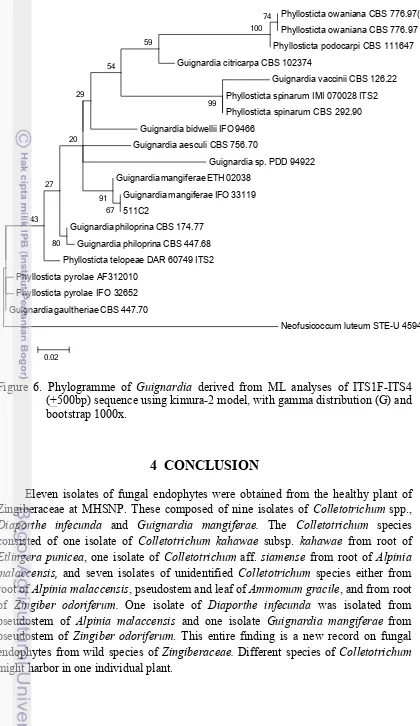
One isolate of Guignardia was obtained from pseudostem of Zingiber odoriferum. A set of reference strain (Table 4) and Neofusicoccum luteum (outgroup) were used in the phylogenetic analyses. Based on phylogenetic study on ITS sequence, the specimen was named Guignardia mangiferae (Figure 6). Confirmation using morphological data cannot be done since this isolate formed sterile fruit bodies. Guignardia mangiferae is closely related to G. citricarpa and G. musae in one clade (Glienke et al. 2011). The difference is G. musae was reported pathogenic in nature (Wulandari et al. 2010). Guignardia citricarpa were also reported vary in genetic both as pathogen or endophyte (Glienke-Blanco et al. 2002) whereas G. mangiferae were found as non-pathogenic (Baayen et al. 2002). Endophytic Guignardia apparently to have wide range of host plant as reported by Okane et al. (2003) that Guignardia endophyllicola were successfully isolated from healthy leaves of 94 plant species from Japan and Thailand. This is the first report on the presence of endophytic Guignardia mangiferae in Indonesia isolated from Zingiberaceous plant.
Table 4 List of reference Guignardia strain used in this study
Species Culture accession number ITS Genbank accession number
Phyllosticta owaniana CBS 776.97(2) CBS 776.97
JF261462 AF312011
P. podocarpi CBS 111647 AF312013
P. spinarum IMI 070028 CBS 292.90
AY042907 AF312009
P. telopae DAR60749 AY042909
P. pyrolae AF312010 IFO 32652
AF312010 AB041242
Guignardia citricarpa CBS 102374 FJ824767
G. vaccinii CBS 126.22 AB095508
G. bidwellii IFO 9466 AB095509
G. aesculi CBS 756.70 AB095504
Guignardia sp. PDD 94922 FJ655899
20
Figure 6. Phylogramme of Guignardia derived from ML analyses of ITS1F-ITS4 (+500bp) sequence using kimura-2 model, with gamma distribution (G) and bootstrap 1000x.
4 CONCLUSION
Eleven isolates of fungal endophytes were obtained from the healthy plant of Zingiberaceae at MHSNP. These composed of nine isolates of Colletotrichum spp., Diaporthe infecunda and Guignardia mangiferae. The Colletotrichum species consisted of one isolate of Colletotrichum kahawae subsp. kahawae from root of Etlingera punicea, one isolate of Colletotrichum aff. siamense from root of Alpinia malaccensis, and seven isolates of unidentified Colletotrichum species either from root of Alpinia malaccensis, pseudostem and leaf of Ammomum gracile, and from root of Zingiber odoriferum. One isolate of Diaporthe infecunda was isolated from pseudostem of Alpinia malaccensis and one isolate Guignardia mangiferae from pseudostem of Zingiber odoriferum. This entire finding is a new record on fungal endophytes from wild species of Zingiberaceae. Different species of Colletotrichum might harbor in one individual plant.
REFERENCES
Arnold AE, Maynard Z, Gilbert GS, Coley PD, Kursar TA. 2000. Are tropical fungal endophytics hyperdiverse? Ecol Lett 3:267-274.
Baayen RP et al. 2002. Nonpathogenic isolates of the citrus black spot fungus, Guignardia citricarpa, identified as a cosmopolitan endophyte of woody plant G. mangiferae (Phyllosticta capitalensis). Phytopathology 92:464-477.
Bussaban B, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD. 2001. Endophytic fungi from Ammomum siamense. Can J Microbiol 47:943-948.
Cannon PF, U Damm, PR Johnston, and BS Weir. 2012. Colletotrichum - current status and future directions. Stud Mycol 73:181-213.
Dhetchuvi JB, Wortley AH, Harris DJ. 2011. A new species of Aframomum (Zingiberaceae) from Central Africa. Phytotaxa 28:31-34.
Freeman S, Katan T. 1997. Identification of Colletotrichum species responsible for anthracnose and root necrosis of strawberry in Israel. Phytopathology 87:516-521.
Gardes M, Bruns TD. 1993. ITS primer with enhanced spesifity for basidiomycetes - application to the identification of mychorrhizae and rusts. Mol Ecol 2:113-118.
Glienke-Blanco C, Aguilar-Vildoso C, Vieira MLC, Barroso PAV, Azevedo JL. 2002. Genetic variability in endophytic fungus Guignardia citricarpa isolated from citrus plants. Genet Mol Biol 25:251-255.
Glienke C et al. 2011. Endophytic and pathogenic Phyllosticta species, with reference to those associated with citrus black spot. Persoonia 26:47-56.
Gomes RR et al. 2013. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31:1-41.
Guo LD, Hyde KD, Liew ECY. 1998. A method to promote sporulation in palm endophytic fungi. Fungal Divers 1:109-113.
Huang WY, Cai YZ, Hyde KD, Corke H, Sun M. 2008. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Divers 33:61-75.
Hyde KD et al. 2010. A case for re-inventory of Australia’s plant pathogens. Persoonia 25: 50-60
Ilyas M, Kanti A, Jamal Y, Hertina, Agusta A. 2009. Biodiversity of endophytic fungi associated with Uncaria gambier Roxb. (Rubiaceae) from West Sumatra. Biodiversitas 10:23-28.
Jeffrey LSH, Son R, Tosiah S. 2008. Preliminary screening of endophytic fungi isolated from medicinal plants at MARDI Sessang, Sarawak for their bioactivity. J Trop Agric Food Sci 36:121-126.
22
Mahadtanapuk S, Sanguansermsri M, Cutler RW, Sardsud V, Anuntalabhochai S. 2007. Control of antrachnose caused by Colletotrichum musae on Curcuma alismatifolia Gagnep. using antagonistic Bacillus spp. Am J Agric Biol Sci 2:54-61.
Manter DK, Vivanco JM. 2007. Use of the ITS primers, ITS1F and ITS4 to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J Microbiol Methods 71:7-14.
Nilsson RH et al. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1:e59.
Noverita, Fitria D, Sinaga E. 2009. Isolasi dan uji aktivitas antibakteri jamur endophytic dari daun dan rimpang Zingiber ottensii Val. J Farmasi Indones 4:171-179.
Okane I, Lumyong S, Nakagiri A, Ito T. 2003. Extensive host range of an endophytic fungus, Guignardia endophyillicola (anamorph: Phyllosticta capitalensis). Mycoscience 44:353-363.
Palarpawar MY, Ghurde VR. 1988. Some new hosts of Colletotrichum curcumae. Indian J Mycol Plant Pathol 18: 220-221.
Photita W, Taylor PWJ, Ford R, Hyde KD, Lumyong S. 2005. Morphological and molecular characterization of Colletotrichum species from herbaceous plants in Thailand. Fungal Divers 18:117-133.
Priyadi H et al. 2010. Five hundred plant species in Gunung Halimun Salak National Park, West Java. A checklist including Sundanese names, distribution and use. Bogor: CIFOR.
Rodriguez RJ, White Jr JF, Arnold AE, Redman RS. 2009. Fungal endophytics: diversity and functional roles. New Phytol 182:314-33.
Schulz B, Boyle C. 2005. The endophytic continuum. Mycol Res 109:661-668.
Silva M, Pereira OL. 2007. First report of Guignardia endophyllicola leaf blight on Cymbidium (Orchidaceae) in Brazil. Aust Plant Dis Notes 2:31-32.
Su YY, Cai L. 2012. Polyphasic characterisation of three new Phyllosticta spp. Persoonia 28:76-84.
Thon MR, Nuckles EM, Takach JE, Vaillancourt LJ. 2002. CPR1: A gene encoding a putative signal peptidase that functions in pathogenicity of Colletotrichum graminicola to maize. Mol Plant Microbe Interact 15:120-128.
Weir BS, Johston PR, Damm U. 2012. The Colletotrichum gloeosporioides species complex. Stud Mycol 73:115-180.
White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. New York: Academic Press. p315-322.
Wu D, Larsen K. 2000. Zingiberaceae. Flora of China 24:322-377.
APPENDIXES
Appendix 1 Maximum Likelihood (ML) of 24 different nucleotide substitution models for Colletotrichum species
Models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. Non-uniformity of evolutionary rates among sites may be modeled by using a discrete Gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (+I). Whenever applicable, estimates of gamma shape parameter and/or the estimated fraction of invariant sites are shown. Assumed or estimated values of transition/transversion bias (R) are shown for each model, as well. The analysis involved 113 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 395 positions in the final dataset. Evolutionary analyses were conducted in MEGA ver. 5.2.2 for Mac.
24
Appendix 2 Maximum Likelihood (ML) of 24 different nucleotide substitution models for Diaporthe species
Model Parameter BIC AICc lnL Invariant Gamma R
TN93+G+I 286 13769 11165 -5295.1 0.536704 0.45742 2.7142
K2+G+I 282 13776 11209 -5321.1 0.54091 0.47393 2.5981
GTR+G+I 289 13782 11150 -5285 0.539538 0.46055 2.7621
T92+G+I 283 13792 11216 -5323.6 0.540182 0.47582 2.5902
HKY+G+I 285 13812 11217 -5322.1 0.540521 0.47846 2.5993
TN93+G 285 13846 11251 -5339.2 n/a 0.18821 2.5799
K2+G 281 13853 11294 -5364.8 n/a 0.19027 2.5452
GTR+G 288 13861 11239 -5330.3 n/a 0.18992 2.5938
T92+G 282 13867 11300 -5366.6 n/a 0.19143 2.5457
HKY+G 284 13884 11299 -5364.1 n/a 0.19052 2.5571
JC+G+I 281 14281 11722 -5579 0.541166 0.49931 0.5
JC+G 280 14352 11803 -5620.1 n/a 0.19279 0.5
K2+I 281 14396 11837 -5636.4 0.645702 n/a 2.3267
T92+I 282 14405 11838 -5635.7 0.645702 n/a 2.335
GTR+I 288 14407 11785 -5603.3 0.645702 n/a 2.2519
TN93+I 285 14407 11812 -5620 0.645702 n/a 2.2764
HKY+I 284 14426 11841 -5635.1 0.645702 n/a 2.3385
JC+I 280 14872 12323 -5880.2 0.645702 n/a 0.5
TN93 284 15843 13257 -6343.5 n/a n/a 2.1427
GTR 287 15852 13239 -6331.2 n/a n/a 2.1448
K2 280 15876 13327 -6382.2 n/a n/a 2.1354
T92 281 15890 13331 -6383.3 n/a n/a 2.1367
HKY 283 15912 13335 -6383.2 n/a n/a 2.1362
JC 279 16331 13791 -6615.3 n/a n/a 0.5
Models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. Non-uniformity of evolutionary rates among sites may be modeled by using a discrete Gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (+I). Whenever applicable, estimates of gamma shape parameter and/or the estimated fraction of invariant sites are shown. Assumed or estimated values of transition/transversion bias (R) are shown for each model, as well. The analysis involved 141 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 477 positions in the final dataset. Evolutionary analyses were conducted in MEGA ver. 5.2.2 for Mac.
Appendix 3 Maximum Likelihood (ML) of 24 different nucleotide substitution models for Guignardia species.
Model Parameter BIC AICc lnL Invariant Gamma R
K2+G 39 2039.3 1791.3 -856.3 n/a 0.28558 2.5466
T92+G 40 2041.3 1787 -853.13 n/a 0.29021 2.5658
K2+G+I 40 2047.1 1792.8 -856.04 0.328263 0.59619 2.5617
T92+G+I 41 2049.2 1788.5 -852.87 0.36506 0.69476 2.5835
HKY+G 42 2058.7 1791.7 -853.43 n/a 0.29164 2.5253
HKY+G+I 43 2066.5 1793.2 -853.15 0.350703 0.65958 2.5453
TN93+G 43 2066.6 1793.4 -853.24 n/a 0.29685 2.519
GTR+G 46 2069.4 1777.1 -842.03 n/a 0.27725 2.3434
TN93+G+I 44 2074.4 1794.8 -852.92 0.394907 0.80424 2.539
GTR+G+I 47 2077.4 1778.8 -841.85 0.269129 0.48594 2.3699
JC+G 38 2090.3 1848.7 -885.99 n/a 0.30132 0.5
K2 38 2092.5 1850.9 -887.08 n/a n/a 2.2074
T92 39 2094.3 1846.4 -883.84 n/a n/a 2.2272
JC+G+I 39 2098.1 1850.2 -885.71 0.385253 0.79119 0.5
K2+I 39 2100.8 1852.9 -887.08 0.00001 n/a 2.2074
T92+I 40 2102.7 1848.4 -883.83 0.00001 n/a 2.2272
HKY 41 2110.8 1850.2 -883.71 n/a n/a 2.2205
TN93 42 2117.3 1850.4 -882.77 n/a n/a 2.2202
HKY+I 42 2119.2 1852.3 -883.71 0.00001 n/a 2.2205
GTR 45 2125.5 1839.6 -874.31 n/a n/a 2.2146
TN93+I 43 2125.7 1852.4 -882.77 0.00001 n/a 2.2202
GTR+I 46 2133.9 1841.6 -874.31 0.00001 n/a 2.2146
JC 37 2139.9 1904.7 -915.01 n/a n/a 0.5
JC+I 38 2148.3 1906.7 -915.01 0.00001 n/a 0.5
Models with the lowest BIC scores (Bayesian Information Criterion) are considered to describe the substitution pattern the best. Non-uniformity of evolutionary rates among sites may be modeled by using a discrete Gamma distribution (+G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (+I). Whenever applicable, estimates of gamma shape parameter and/or the estimated fraction of invariant sites are shown. Assumed or estimated values of transition/transversion bias (R) are shown for each model, as well. The analysis involved 20 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 218 positions in the final dataset. Evolutionary analyses were conducted in MEGA ver. 5.2.2 for Mac.
26
BIOGRAPHY
Nicho Nurdebyandaru was born in Jember, East Java Province and graduated in 2004 from SMAN 1 Jember. He entered Institut Pertanian Bogor/IPB (Bogor Agricultural University) in Biology major of undergraduate program. In his second year, he had begun teaching students as a laboratory assistant for practical work until the end of the semester. His research theme was in Microbiology field about the biocontrol bacteria against aphids. That research was funded by DIKTI (Directorate General of Higher Education) via Student Creativity Program (PKMI) and awarded as 3rd best presenter in National Competition. After graduated in December 2008, he worked for multinational dairy company, PT Frisian Flag Indonesia from June 2009 until June 2011 as Microbiologist. He learned and earned many experience on system (ISO, food quality and laboratory management).