MOLECULAR CHARACTERIZATION OF
BEGOMOVIRUS
INFECTING
YARD LONG BEAN (
Vigna unguiculata
subsp.
sesquipedalis
L.) AND
CONSTRUCTION OF ITS SPECIFIC PRIMERS
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
2014
AUTHOR’S STATEMENT
ON THESIS AND ITS SOURCE OF
INFORMATION AND DELEGATION OF COPYRIGHT
I declare that this thesis entitled Molecular Characterization of Begomovirus
Infecting Yard Long Bean (Vigna unguiculata subsp. sesquipedalis L.) and
Construction of Its Specific Primers is my own and authentic work under supervision of a thesis commitee and it is not yet submitted to any universities for any degree fulfillment. Source of information, both published or unpublished by it authors, used for quotations in this thesis is already cited appropriately and
present in thesis‟s Literature Cited chapter.
I hereby delegate my thesis copyright to the Bogor Agricultural University. Bogor, August 2014
Sari Nurulita
Student ID number: A352110081
Copyright delegation of scientific paper from research collaboration with other
SUMMARY
SARI NURULITA. Molecular Characterization of Begomovirus Infecting Yard
Long Bean (Vigna unguiculata subsp. sesquipedalis L.) and Construction of Its Specific Primers. Supervised by SRI HENDRASTUTI HIDAYAT and KIKIN HAMZAH MUTAQIN.
Begomovirus is an important plant viral pathogen group causing serious diseases in several tropical and sub-tropical countries. Infection of Begomovirus
was reported to cause economic losses up to million dollars. Infection of
Begomovirus on horticultural crops in Indonesia was first reported on chilli pepper in 1999 with disease incidence reached 30 to 100%. Infection of Begomovirus on chili pepper is obviously recognized by its typical symptoms involving leaf curling and yellowing.
Yellow mosaic disease outbreaks have also been reported on yard long bean (Vigna unguiculata subsp. sesquipedalis L.) growing area in Java since 2008. At
the same time, similar yellow mosaic disease was first reported in Pakistan and Nepal on Leguminosae. The causal agent of yellow mosaic disease in South Asia was identified as a member of Begomovirus. Yellow mosaic disease on yard long
bean in Java was considered to be caused by Begomovirus.
Begomovirus is the largest genus of the Family Geminiviridae. Molecular characteristics of Begomovirus genome is very unique, comprising twin circular particles, each particle is consisting of single stranded DNA (2.6 – 2.7 kb), bipartite or monopartite. DNA-A and DNA-B of bipartite Begomovirus have
different genome organization and function except for their common region. On the other hand, monopartite Begomovirus has all combination function of DNA-A
and DNA-B in one genome. There are three unique characteristics of common region which is observed on every Begomovirus species, i.e. TATA box, repeated motives (iterons), and hairpin-loop structure.
Begomovirus is only transmitted in nature by insect vector, B. tabaci
(Hemiptera: Aleyrodidae). Studies on virus – vector relationship indicates that
Begomovirus is transmitted in circulative persistent manner. Begomovirus can not be transmitted by mechanical nor seed manner. The nature of Begomovirus of
being insect transmissable and having a wide host range may contribute to its potential to cause high disease incidence and being difficult to control.
Common method for detection of Begomovirus is based on polymerase
chain reaction (PCR). PAL1v1978/PAR1c715 and AV494/AC1048 primer pairs have been reported to work well for different species of Begomovirus. The former pair amplifies common region (CR), whereas the latter amplifies coat protein gene of Begomovirus. PAL1v1978/PAR1c715 was successfully used to detect several
geminivirus from Solanaceae, Leguminosae, Euphorbiaceae, and Malvaceae plants.
The objective of this research is to determine the importance of
Begomovirus infection on yard long bean, to identify species of Begomovirus on
yard long bean and to analyse its molecular character, and to design specific primers for detection of Begomovirus species infecting yard long bean.
Klaten), Jogyakarta (Sleman and Klaten), and West Java (Bogor and Subang) provinces; (ii) virus detection using I-ELISA, PCR, cloning, and sequencing; (iii) molecular characterization of Begomovirus using software BioEdit v.7.0.5, CLC Sequence Viewer, and MEGA 6.06; (iv) construction and use of specific primer;
and (v) Koch‟s Postulate trial using whitefly transmission.
Yellow mosaic disease was found in almost all fields surveyed. Infection of
Potyvirus and Begomovirus was detected using I-ELISA and PCR, respectively.
Both viruses were detected as either single or mixed infection. Begomovirus
infections were detected more often than Potyvirus, which indicated that Begomovirus is predominant in all locations. Begomovirus infecting yard long
bean in all samples has a bipartite genome based on detection using PAL1v1978/PAR1c715 universal primer pair and PBL1v2040/PCR1c, i.e. possessing DNA-A and DNA-B.
Common region sequence of DNA-A has identity >85% to that ofMungbean yellow mosaic India virus (MYMIV). Further phylogenetic analysis to study their relationship showed that all Begomoviruses infecting mungbean (MYMIV and
MYMV) are belong to similar cluster and they are separated from Begomoviruses
which have been reported previously. All MYMIV isolates from Java have three unique characteristics, i.e. three repetitive sequences ATCGGTGT and one invert sequence ACACCGAT, TATA box sequence, and hairpin loop structure sequence which consist of 32 nucleotides GGGCACTCAGCTATA ATATTACCTGAGTGCCC. All CR components of all MYMIV isolates from Java has similarity with MYMIV from India, Pakistan, and Nepal with >85% homology; whereas homology among Java isolates reached >90 – 100%. Spesific primer pairs MYF/MYR and MY1/MY2 were successfully amplified specific DNA target of 1000 bp and 238 bp, respectively and can be used to distinguish MYMIV from other Begomoviruses.
Transmission study to fulfill Koch‟s Postulateusing three yard long bean
varieties resulted in similar yellow mosaic symptom with those in the field.
„Parade‟ is the most susceptible variety with 100% (5/5) disease incidence, followed by „New Jaliteng‟ and „Wulung‟ with both caused 40% (2/5) disease incidence. Incubation period of Begomovirus in all varieties was 14 days in
average.
Diagnosis based on molecular detection and transmission study confirmed that Mungbean yellow mosaic India begomovirus is the main causal agent of yellow mosaic disease of yard long bean. Disease control strategy to suppress incidence, severity and dissemination of the disease should be undertaken promptly.
RINGKASAN
SARI NURULITA. Karakterisasi Molekuler Begomovirus yang Menginfeksi
Kacang Panjang (Vigna unguiculata subsp. sesquipedalis L.) dan Pembuatan Primer Spesifik. Dibimbing oleh SRI HENDRASTUTI HIDAYAT dan KIKIN HAMZAH MUTAQIN.
Begomovirus merupakan salah satu patogen penting yang menginfeksi beberapa komoditas hortikultura utama di negara tropis dan sub-tropis. Kerugian ekonomi akibat infeksi Begomovirus di beberapa negara dilaporkan mencapai
jutaan dolar. Infeksi Begomovirus di Indonesia pada tanaman hortikultura pertama kali dilaporkan tahun 1999 pada tanaman cabai dengan kejadian penyakit 30 –
100%. Tanaman cabai yang terinfeksi Begomovirus mempunyai gejala yang khas,
yaitu daunnya mengeriting dan menguning.
Gejala mosaik kuning juga mulai ditemukan di sejumlah pertanaman kacang panjang pada tahun 2008 dan kejadian penyakit semakin meningkat terutama di beberapa daerah di Jawa. Gejala yang sama juga dilaporkan pada tanaman
Leguminosae di India, Bangladesh, Pakistan, dan Nepal. Penyakit mosaik kuning di kawasan Asia Selatan ini disebabkan oleh infeksi Begomovirus. Penyakit
mosaik kuning di Jawa diduga juga disebabkan oleh Begomovirus.
Begomovirus merupakan salah satu genus terbesar dari famili Geminiviridae. Begomovirus mempunyai karakter molekuler yang unik dengan partikel kembar (bipartit) atau tunggal (monopartit). Partikel bipartit Begomovirus
terdiri dari DNA-A dan DNA-B, masing-masing berukuran 2.6 – 2.7 kb dengan fungsi yang berbeda. Partikel monopartit mempunyai fungsi gabungan dari DNA-A dan DNDNA-A-B. Identitas setiap spesies Begomovirus ditemukan pada bagian common region (CR). Setiap CR Begomovirus mempunyai tiga komponen unik,
yaitu sekuen berulang (iteron), sekuen TATA box, dan struktur hair pin loop. Penularan Begomovirus di lapangan sebagian besar melalui vektornya
Bemisia tabaci Gen. (Hemiptera: Aleyrodidae) secara persisten sirkulatif. Begomovirus tidak dapat ditularkan baik secara mekanis maupun benih. Kisaran inang yang luas dan penyebaran melalui vektor membuat kejadian penyakit
Begomovirus tinggi dan sulit dikendalikan.
Deteksi Begomovirus umumnya dilakukan dengan metode Polymerase chain
reaction (PCR). Metode PCR merupakan salah satu teknik deteksi yang sensitif dan cepat. Primer merupakan salah satu komponen utama dalam proses PCR. PAL1v1978/ PAR1c715 dan AV494/ AC1048 merupakan primer universal yang banyak digunakan untuk mendeteksi Begomovirus. Primer PAL1v1978/ PAR1c715 dilaporkan dapat mendeteksi Begomovirus yang menginfeksi beberapa tanaman dari famili Solanaceae, Leguminosae, Euphorbiaceae, dan Malvaceae.
Pasangan primer PAL1v1978/ PAR1c715 mengamplifikasi bagian CR, sedangkan primer AV494/ AC1048 mengamplifikasi pada bagian protein selubung.
Penelitian ini bertujuan untuk melaporkan pentingnya infeksi Begomovirus
pada tanaman kacang panjang, mengidentifikasi spesies Begomovirus pada
tanaman kacang panjang, menganalisis karakter molekuler dan membuat primer spesifik untuk mendeteksi spesies Begomovirus yang menginfeksi tanaman
Penelitian ini dilakukan dengan beberapa tahapan, yaitu: (i) survei penyakit di beberapa lokasi pertanaman kacang panjang di provinsi Jawa Tengah (Tegal, Magelang, dan Klaten), D.I. Yogyakarta (Sleman dan Kalasan), serta Jawa Barat (Bogor dan Subang); (ii) deteksi beberapa virus yang menginfeksi tanaman kacang panjang menggunakan I-ELISA serta deteksi Begomovirus menggunakan PCR, cloning, dan sekuensing; (iii) analisis karakter molekuler Begomovirus
menggunakan perangkat lunak BioEdit v.7.0.5, CLC Sequence Viewer, dan
MEGA 6.06; (iv) membuat primer spesifik menggunakan perangkat lunak
Oligonucleotide Calculator dan PrimerBLAST; serta (v) uji postulat Koch
menggunakan penularan kutukebul dan inokulum dari lapangan untuk membuktikan agen penyebab gejala mosaik kuning.
Gejala mosaik kuning ditemukan hampir di semua lokasi survei. Berdasarkan hasil deteksi menggunakan I-ELISA dan PCR ditemukan adanya infeksi Potyvirus tunggal, Begomovirus tunggal, serta campuran antara
Begomovirus dan Potyvirus. Hasil deteksi seluruh sampel menunjukkan dominasi infeksi Begomovirus di beberapa lokasi. Hasil deteksi menggunakan primer
universal PAL1v1978/ PAR1c715 dan PBL1v2040/ PCR1c membuktikan bahwa
Begomovirus yang menginfeksi tanaman kacang panjang di Indonesia memiliki genom bipartite, yaitu memiliki DNA-A dan DNA-B.
Sekuen bagian CR DNA-A isolat Begomovirus hasil survei mempunyai nilai
kemiripan lebih dari 85% dengan spesies Mungbean yellow mosaic India virus
(MYMIV). Semua isolat MYMIV asal Jawa pada analisis filogenetik mengelompok bersama isolat MYMIV asal India, Banglades, Nepal, dan Pakistan. Isolat MYMIV ini terpisah dari spesies-spesies Begomovirus Indonesia yang telah
dilaporkan sebelumnya.
Semua isolat MYMIV asal Jawa memiliki tiga karakter unik CR, yaitu tiga sekuen berulang ATCGGTGT dan satu invert sequence ACACCGAT, sekuen TATA box, dan struktur pembentuk hairpin loop yang terdiri dari 32 sekuen nukleotida GGGCACTCAGCTATAATATTACCTGAGTGCCC. Semua komponen CR isolat MYMIV asal Jawa memiliki kesamaan dengan CR isolat MYMIV asal India, Pakistan, dan Nepal. Sekuen CR semua isolat MYMIV asal Jawa juga memiliki nilai homologi >85% dengan isolat MYMIV asal Banglades, India, Nepal, dan Pakistan serta memiliki homologi >90 – 100% antara sesama isolat asal Indonesia. Primer spesifik MYF/ MYR dan MY1/ MY2 yang didesain dalam penelitian ini berhasil mengamplifikasi MYMIV secara spesifik berturut-turut pada 1000 bp dan 238 bp.
Hasil penularan (uji postulat Koch) menggunakan kutukebul B. tabaci pada tiga varietas kacang panjang menghasilkan gejala mosaik kuning sama dengan
yang ada di lapangan. Varietas „Parade‟ merupakan varietas paling rentan dengan kejadian penyakit 100% (5/5) diikuti oleh „New Jaliteng‟ dan „Wulung‟ masing -masing sebesar 40% (2/5). Masa inkubasi Begomovirus pada semua varietas adalah 14 hari.
Diagnosis penyebab penyakit mosaik kuning kacang panjang berdasarkan deteksi molekuler dan uji penularan membuktikan Mungbean yellow mosaic India begomovirus sebagai agens utama penyebab penyakit. Strategi pengendalian untuk menekan kejadian, keparahan dan penyebaran penyakit harus segera diupayakan.
©2014, Bogor Agricultural University
All Right Reserved
It is prohibited to make any quotations from part or whole of this thesis without citing the author or the copyright holder. Quotation is allowed as long as for education, research, scientific writing, scientific report, research proposal or scientific review purposes only; those quotations should not produce any adverse effects to Bogor Agricultural University.
Thesis
in partial fulfillment of the requirements for the degree of Master of Science
at the
Phytopathology Study Program
MOLECULAR CHARACTERIZATION OF
BEGOMOVIRUS
INFECTING
YARD LONG BEAN (
Vigna unguiculata
subsp.
sesquipedalis
L.) AND
CONSTRUCTION OF ITS SPECIFIC PRIMERS
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY BOGOR
2014
Thesis title : Molecular Characterization of Begomovirus Infecting Yard Long Bean (Vigna unguiculata subsp. sesquipedalis L.) and Construction of Its Specific Primers
Name : Sari Nurulita SIN : A352110081
Approved by Committee members
Prof Sri Hendrastuti Hidayat, PhD Supervisor
Kikin Hamzah Mutaqin, PhD Co-Supervisor
Acknowledged by
Head of Phytopathology Study Program
Prof Sri Hendrastuti Hidayat, PhD
Dean of Graduate School
Dr Ir Dahrul Syah, MScAgr
PREFACE
I am very grateful to Allah Subhanallahuwataala for the mercy, so I can
finish this thesis. From this moment I would also like to thank the people who specially contribute to my education.
I would like to express my thankfulness to my committee members, Prof. Sri Hendrastuti Hidayat and Dr. Kikin Hamzah Mutaqin for their guidance and support, both in scientific and personal matters. Their patience and excellent advice made this work enjoyable and rewarding. I am very thankful to Dr. Gede Suastika for his suggestions during thesis defense.
I am very thankful to Dr. John Thomas and Matthew Webb, Department of Agriculture, Forestry and Fisheries (DAFF) Queensland – Australia, for their technical assistance especially for teaching me laboratory technique and providing molecular material in Plant Virology Laboratory, Australia. I also thank Dr. Cherie Gambley for the guidance on methodology to design pecific primers. I thank Tuti Legiastuti, SSi for her assistance in Plant Virology Laborator, IPB. I would also like to thank Dr. Sri Sulandari, Gadjah Mada University, for helping me during the survey.
I am very thankful to AusAID Funded Economic Cooperation Work Program of the ASEAN – Australia – New Zealand Free Trade Agreement for funding my research. I am very thankful to I-MHERE B.2C IPB for granting me a master scholarship.
I would also like to thank my classmates “Phytopathology class 2011”,
Plant Virology Laboratory members, especially Dwiwiyati Nurul Septariani for their supports and cooperations. My appreciation goes to my family who always support and care me from far away.
Partial of this thesis has been presented in International Workshop on Collaborative Educations for Sustainability of Agriculture and Environment, Ibaraki University – Japan 2012; Indonesian Phytopathology Society, Padang 2013; also has been submitted to Biotropia Journal – SEAMEO Biotrop.
Bogor, August 2014
TABLE OF CONTENTS
LIST OF TABLES vi
LIST OF FIGURES vi
LIST OF APPENDIX vi
INTRODUCTION 1
Background 1
Hypothesis 3
Research Objectives 3
Research Advantages 3
LITERATURE REVIEWS 5
Importance of Begomovirus Infection on Agricultural Production 5 Disease Control Strategy for Begomovirus Infection 5
Legume – Infecting Begomovirus 6
Biological Characteristics of Begomovirus 7
Organization and Molecular Characteristics of Begomovirus 8
Diagnosis and Detection of Begomovirus 9
MATERIALS AND METHODS 11
Field Survey and Samples Collection 11
Detection and Identification of Viruses from Infected Leaves 11
Virus Detection by I-ELISA 11
Begomovirus Detection by PCR 12
Cloning Strategy of DNA-A of Begomovirus 12
DNA Sequencing 14
Construction of Specific Primers 14
Evaluation of Primer Specificity 16
Koch's Postulate 16
RESULTS AND DISCUSSION 17
Identification of Begomovirus from Field Samples 17
Cloning of DNA-A Begomovirus 19
Sequence Identity Analysis of Begomovirus Infecting Yard Long
Bean 20
Analysis of Common Region of Begomovirus Infecting Yard Long
Bean 21
Specificity of MYMIV Specific Primers 27
Koch‟s Postulate 28
General Discussion 29
CONCLUSION 31
APPENDIXES 40
LIST OF TABLES
1 Genome organization of Begomovirus 8
2 Degenerate primers for amplification of Begomovirus using polymerase
chain reaction 12
3 Specific primers for amplification of Begomovirus infecting yard long
bean 15
4 Detection of CMV, Potyvirus, and Begomovirus from leaf samples
collected from various locations using I-ELISA and PCR 19 5 Nucleotide sequence homology (%) of Begomovirus infecting yard long
bean in Java with other Begomoviruses reported earlier in GenBank 20
6 Comparison of repetitive sequences (iteron) between MYMIV and
MYMV on common region 24
7 Nucleotide sequence homologies (% identity) of common region among
Begomovirus 26
LIST OF FIGURES
1 Research flow chart: “Molecular Characterization of Begomovirus
Infecting Yard Long Bean (Vigna unguiculata subsp. sesquipedalis L.)
and Construction of Its Specific Primers” 4
2 Genome organization of Begomovirus: bipartite (above) and
monopartite (below) 9
3 Hair pin loop structure of Begomovirus 10
4 pCR®4-TOPO® vector for cloning 13
5 Alignment of conserved region to design specific primer for
Begomovirus infecting yard long beans 15 6 Construction of specific primer using primer BLAST program at NCBI 16 7 Various symptoms found associated with yellow mosaic disease on
yard long bean 17
8 Visualization of PCR products of Begomovirus amplification from leaf
samples using universal primers PAL1v1978/ PAR1c715 on 1 %
agarose gel 18
9 Visualization of PCR products of Begomovirus amplification from leaf samples using universal primers PBL1v2040/ PCR1c on 1 % agarose
gel 18
10 Visualization of PCR product from Begomovirus clone(pTgl) 20 11 Phylogenetic analysis based on alignment of partial nucleotide
sequences of the DNA-A of Begomoviruses using Mega 6.06
(Algorithm Neighbour Joining with 1000 bootstraps replicates) 21 12 Alignment of nucleotide sequences of the CR of Mungbean yellow
mosaic India virus (MVMIV) isolates from Java with other reported
MYMIV in Genbank 22
13 Hairpin loop structures of several Begomovirus isolates 25 14 Amplification of partial DNA-A Begomovirus genome using universal
primer PAL1v1978/PAR1c715 (A), specific primers MYF/MYR (B),
LIST OF APPENDIXES
1 Sequence alignment of all MYMIV isolates 40
INTRODUCTION
Background
Begomovirus is an important plant viral pathogen group causing serious diseases in several tropical and sub-tropical countries (Mansoor et al. 2003;
Varma and Malathi 2003). Infection of Begomovirus resulted in crop damage and
yield loss up to 20% or equal to US$ 140 million economic losses in tomato production in Florida, US (Moffat 1999; Polston et al. 1999). Mosaic disease of
cassava in East and Central Africa was also the reason for US$ 1.9 – 2.7 billion yield loss (Patil and Fauquet 2009). Similar situation was reported from cucurbit production in India which was suffered from yellow mosaic disease (Varma and Malathi 2003; Varma et al. 1992). In Indonesia, serious infection of Begomovirus
caused disease epidemic on some chilli production regions in Central Java and West Java Province, and Special District of Jogyakarta (Sulandari et al. 2006). Infection of Begomovirus in chilli pepper produced 30-100% disease incidence
since 1999 (Hidayat et al. 2006).
Yellow mosaic disease outbreak has been reported on yard long bean (Vigna unguiculata subsp. sesquipedalis L.) growing area in Java since 2008. Bean common mosaic virus (BCMV) and Cucumber mosaic virus (CMV) were
identified from leaf samples showing yellow mosaic symptom collected from yard long bean growing area in Java (Damayanti et al. 2009). In the meantime similar yellow mosaic disease was first reported from Pakistan and Nepal on grain legume, mungbean, cowpea, soybean, kidney bean and Rhynchosia capitata.
Further identification showed the involvement of bipartite Begomovirus on this disease (Ilyas et al. 2010; Shahid et al. 2012).
Begomovirus is the largest genus of the Family Geminiviridae. Its natural
spread relies on its specific insect vector, Bemicia tabaci (Hemiptera : Aleyrodidae) which transmit the virus in circulative persistent manner. Most
Begomovirus is not transovarially transmitted, except TYLCV from Israel
(Ghanin et al. 1998). Since B. tabaci is a polyphagous insect, Begomovirus has a high chance to infect many plant species. For instance, tomato leaf curl disease (ToLCD) caused by Begomovirus has been observed on many countries. Tomato leaf curl New Delhi virus (ToLCNDV) was investigated as new emerging virus
causing yellow mosaic disease on eggplant in India (Pratap et al. 2011). ToLCNDV was also reported infecting cucumber plant in Central Java, Indonesia (Mizutani et al. 2011). Another ToLCD caused by Tomato leaf deformation virus
was characterized as a new world monopartite Begomovirus infecting tomato in Peru and Ecuador (Melgarejo et al. 2013). A new Begomovirus associated with tomato yellow leaf curl and eggplant yellow mosaic disease, Tomato yellow leaf curl Kanchanaburi virus was found in Thailand (Green et al. 2003), Indonesia (Nurul and Tsai 2009), and Laos (Tang et al. 2014). Three Begomoviruses were identified associated with bean golden mosaic disease in Nicaragua i.e., Bean golden yellow moaic virus (BGYMV), Squash yellow mild mottle virus
2
Molecular characteristics of Begomovirus genome is very unique; it posses twin particles, each consisting of single stranded DNA (2.6 – 2.7 kb), bipartite or monopartite (Hull 2002). DNA-A and DNA-B of Begomovirus have different
genome organization and function except their common region (CR). Among other important function encodes by genome of DNA-A are virus replication and insect transmission, whereas those encodes by DNA-B are systemic infection and symptom development. Common region of both DNA-A and DNA-B each consists of about 200 nucleotides and its sequence is highly identical for one species of Begomovirus. In contrast, common region of each species of Begomovirus is distinctinve, except for loop region on hairpin structure, i.e. 5‟-
TAATATTAC -3‟ which is known as the conserved nona-nucleotide region (Lazarowitz 1987). There are three unique characteristics of common region which is observed on every Begomovirus species, i.e. TATA box, repeated motifs
(iterons), and hairpin-loop structure (Hidayat et al. 2008; Lazarowitz 1987; Trisno
et al. 2009). Begomovirus possesing >92% similarity on their hairpin loop structure can be grouped as one species (Fauquet et al. 2005).
The most common method for virus detection is using serological-based technique, but different situation occurred for Begomovirus. Cross reaction and low specificity are the reasons why serological method is not preferred for detection of Begomovirus (Kushwaha et al. 2010). Method for detection of Begomovirus is commonly based on polymerase chain reaction (PCR) technique using universal primers. Several universal primers has been reported to work well for different species of Begomovirus. Rojas et al. (1993) used degenerate primer
PAL1v1978/ PAR1c715 to detect several geminivirus from infected plants belong to Solanaceae, Leguminosae, Euphorbiaceae, and Malvaceae from South America. Li et al. (2004) used a pair of degenerate primer SPG1/ SPG2 to detect
geminivirus infecting sweet potato and other Ipomoea sp. Another pair of degenerate primer, AV494/ AC1048, was used to detect coat protein of subgroup III geminivirus (Wyatt and Brown 1996).
Despite the benefit of using universal primers, specific primers might increase efficiency and effectiveness of detection method. Specific primer is used to positively identify and determine the species involved (Ye et al. 2012).
Torres-Pacheco et al. (1996) used three sets of specific primers to detect intergenic region
(IR) of DNA-A of Pepper jalapeño virus (PJV)/ Texas pepper geminivirus (TPV), IR of DNA-B Chino del tomato virus (CdTV), and IR of DNA-A Pepper huasteco virus (PHV) which infecting pepper in Mexico. Pratap et al. (2011) developed
specific primers to confirm agroinfectious clone of DNA-A and DNA-B ToLCNDV infecting eggplant. Primer pair of HOG1/ HOG2 was specifically designed to amplify full length DNA-A MYMIV using rolling circle replication method (Shahid et al. 2012). Availability of specific primer for specific virus will
be very useful for early diagnosis especially when immediate control strategy is required to take place.
3 Hypothesis
(i) There are multiple viruses associated with yellow mosaic disease on yard long bean, one of them is Begomovirus.
(ii) Begomovirus infecting yard long bean is different from those previously reported in Indonesia.
(iii) Begomovirus infecting yard long bean in Indonesia has close genetic
relationship with other Begomoviruses infecting Leguminosae in Asia.
(iv) Specific primer for Begomovirus infecting yard long bean will specifically amplify virus target.
Research Objectives
Objective of this research is to determine the importance of Begomovirus
infection on yard long bean, to identify species of Begomovirus on yard long bean
and to analyse its molecular character, and to design specific primers for detection of Begomovirus species infecting yard long bean.
Research Advantages
4
Field Survey and Virus Detection West Java Province (Bogor and Subang); Central Java Province (Tegal, Klaten, and Magelang);
Special District of Jogyakarta (Bantul and Sleman)
: process : future process
: result : future prospective result
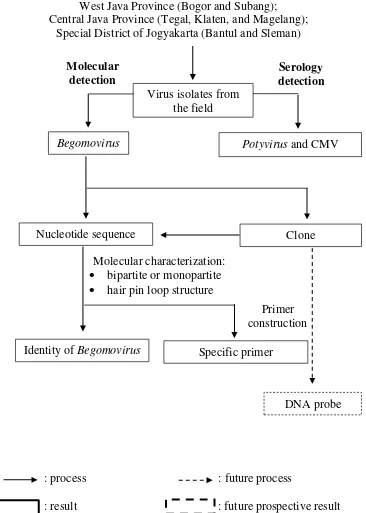
Fig 1 Research flow chart: “Molecular Characterization of Begomovirus Infecting Yard Long Bean (Vigna unguiculata subsp. sesquipedalis L.) and
Construction of Its Specific Primers”
Virus isolates from the field
Begomovirus Potyvirus and CMV
Nucleotide sequence
Specific primer Primer construction Identity of Begomovirus
Clone
DNA probe Molecular
detection
Serology detection
Molecular characterization:
bipartite or monopartite
5
LITERATURE REVIEWS
Importance of Begomovirus Infection on Agricultural Production
Begomovirus was reported to cause serious problems in the sub-tropical
and tropical region and its infection may potentially cause yield lost (Varma et al.
2011). Tomato production in Florida, USA was suffered from tomato yellow leaf curl disease (TYLCD) which caused 20% yield lost or US$ 140 million economy losses (Moffat 1999; Polston et al. 1999). TYLCD was also reported causing yield
loss up to 99% of tomato production in Central America, Europe, Middle East, and South East Asia (Polston and Anderston 1993; Rochester et al. 1994). Legume-infecting Begomovirus, Bean golden mosaic virus, caused 20 – 95% and
36.7% disease incidence in Nicaragua and Argentina, respectively (Karkashian et al. 2011; Alemandri et al. 2012).
Similar condition was also reported from India and Africa, in which up to 50% yield loss of cucurbit in Northern India was occurred in 2001 due to Tomato leaf curl virus (ToLCV) infection (Varma and Malathi 2003). Disease incidence caused by Tomato leaf curl New Delhi virus (ToLCNDV) reached 60 – 66% and
damaged eggplant production in India (Pratap et al. 2011). Serious infection of Begomovirus on cassava caused economy losses of US$ 1.9 – 2.7 billion in East and Central Africa (Patil and Fauquet 2009). Yellow mosaic disease on legume was a very serious problem in India because it caused up to US$ 300 million (Varma et al. 1992). MYMV infection in India ranged from 4 to 40% depend on
crop variety and management (Bashir et al. 2006). Another species, MYMIV, was reported infecting Phaseolus vulgaris in Pakistan (Naimuddin et al. 2011) and Vigna mungo in Nepal (Shahid et al. 2011) with disease incidence reached 70 –
100% and 70 – 80%, respectively.
Incidence of Begomovirus was also reported as serious problem in Indonesia.
Pepper yellow leaf curl Indonesia virus (PepYLCIV) was the main disease
problem on chili pepper in Central Java, Yogyakarta, and West Java with 70 –
100% and 10 – 35% disease incidence for Capsicum frutescence and C. annuum, respectively (Sulandari et al. 2006). Tomato leaf curl virus (ToLCV) infected
tomato in Java and Sulawesi with 100% disease incidence (Aidawati et al. 2005;
Tsai et al. 2009). Infection of Tobacco leaf curl virus Begomovirus on tobacco in East Java reached 30% disease incidence (Trisusilowati 1990).
Disease control strategy for Begomovirus infection
Various control methods has been applied and evaluated in order to suppress disease incidence caused by Begomovirus. Most of the control strategy recommended was based on insect vector management. Preventive strategy in the seedling and young plant using seedbeds with plastic polyethylene sheets was common and was able to avoid Begomovirus transmission by whitefly (Hidayat et al. 2010). Utilization of UV-absorbing plastic films as barrier and neem extract can control whitefly population in the greenhouse (Antignus 1996; Senguttuvan et al. 2005). Combination of screen and insecticide sprays in the greenhouse can
6
Cultural practice using border plants, such as maize, yard long bean, and
Crotalaria sp. as physical barrier around chili pepper plantation can delayed PYLCIV for the first 30 days after transplanting (Hidayat et al. 2010). The maize
and cowpea intercrops on cassava plantation in southern Cameroon can reduce B. tabaci population and cassava mosaic disease incidence up to 50% and 20%, respectively (Fondong et al. 2002).
Treatment using bio-control agent such as PGPR (plant growth promoting rhizobacteria) or PGPF (plant growth promoting fungi) was reported on several studies in attempt to control Begomovirus infection. These treatments was
potentially delayed incubation period and decreased disease incidence of PepYLCIV (Hidayat et al. 2010), induced systemic resistance of PepYLCIV (Damayanti 2013) and CMV (Raupach et al. 1996), and reduced disease incidence of ToMoV up to 40 days after treatment (Murphy et al. 2000).
Growing resistance varieties has been recommended as control strategy for
Begomovirus (Mason et al. 2000) and effort to develop resistance varieties has been studied for various crops. Screening of germplasms from various sources is important to identify source of resistance in the breeding program (Larsen and Porter 2011). Evaluation of mungbean breeding lines was reported but all germplasms was susceptible against MYMV based on observation on incubation period and disease incidence (Shad et al. 2006). Recently, Akhtar et al. (2011)
reported there were 35 genotypes of mungbean showing moderate resistant toward MMIV infection. Santoso (2008) reported F1-TYLCV which was crossed from AVRDC germplasm and commercial seed (F1 FLA456/ Intan and FLA456/ CL6046) showed resistant response up to 68% and 66% toward TYLCV infection, respectively.
The use of transgenic lines to reduce Begomovirus infection were reported.
RNA interference mechanism (RNAi/ silencing mechanism) can reduce viral DNA replication of African cassava mosaic virus (ACMV) up to 99% (Vanitharani et al. 2003). Transgenic cassava plant which resistant to cassava
mosaic disease was also observed by Zhang et al. (2005). Another mechanism,
intron-spliced hairpin construction was used to delay seven weeks of TYLCV incubation period in tomato transgenic line (Zrachya et al. 2007) and recovery
MYMIV infection in blackgram (Vigna mungo) (Pooggin et al. 2003).
Legume–Infecting Begomovirus
Legume-infecting Begomovirus was reported as serious problem in sub-tropical and sub-tropical countries (Varma and Malathi 2003). Symptom variation is reported although it can be differentiated into two types, i.e. golden mosaic and yellow mosaic. Golden mosaic disease was generally found in the new world such as South and Central America and the Caribbean Basin. Bean golden mosaic virus
(BGMV) and Bean golden yellow mosaic virus (BGYMV) have been identified as
causal agent of bean golden mosaic disease in Brazil, Puerto Rico, and Nicaragua (Gilbertson et al. 1993; Karkashian et al. 2011). Bean golden mosaic disease is recognized by its typical symptoms including bright golden color of the leaves, stunting, and smaller pods.
7 on legume characterized by bright yellow leaves with green spot or complete yellowing, smaller pods and seeds was first reported from India (Nariani 1960). Similar symptom was also reported in other Asian countries, such as Pakistan (Qazi et al. 2007), Nepal (Shahid et al. 2012), and Thailand (Honda et al. 1983). There are five species of Begomovirus causing yellow mosaic disease in Asia, i.e.
Mungbean yellow mosaic virus (MYMV), Mungbean yellow mosaic India virus
(MYMIV), Horsegram yellow mosaic virus (HgYMV), Dolichos yellow mosaic virus (DoYMV) (Maruthi et al. 2006; Qazi et al. 2007), and a new species proposed as Rhynchosia yellow mosaic virus (RhYMV) (Ilyas et al. 2009). These
species were distinct to other Begomovirus from new world which was identified
as causal agent of yellow mosaic disease on Leguminosae (Briddon and Stanley 2006).
MYMV and MYMIV were reported as the most destructive agent for some Leguminosae crops in India and Pakistan. MYMV was found to infect Leguminosae in Southern and Western India (Morinaga et al. 1993) whereas MYMIV was reported to infect Leguminosae in Northern and Central region (Mandal et al. 1997), and in Pakistan (Ilyas et al. 2010). MYMV and MYMIV
have close relationship based on phylogenetic analysis except in common region (CR). CR of MYMIV has >95% identity with the same isolate but only <75%
Regenmortel 2000). Mastrevirus has a monopartite genome consists of single circular DNA, infecting monocotyledonous, transmitted by leafhopper Cicadulina mbiia (Hemiptera: Cicadelliadae), its member type is Maize streak virus. Topocuvirus, named after its member Tomato pseudo-curly top virus, has also a
monopartite genome, but infecting dicotyledonous, and transmitted by treehopper
Micrutalis malleifera (Hemiptera: Cicadellidae). Curtovirus, its member type is Beet curly top virus, has similar genome organization and host with Topocuvirus
but it is transmitted by leafhopper Circulifer tenellus (Hemiptera: Cicadellidae).
Begomovirus, named after its member Bean golden mosaic virus, is the largest group of Geminiviridae, having twin circular particles, each consisting of single
stranded DNA (2.6 – 2.7 kb), bipartite, infecting dicotyledonous, and transmitted by whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) (Hull 2002).
Begomovirus is only transmitted in nature by its insect vector, B. tabaci.
Studies on virus – vector relationship indicated that Begomovirus is transmitted in
circulative persistent manner and not transovarial, except for TYLCV-Israel (Ghanim et al. 1998). Furthermore, Aidawati et al. (2002), Sulandari (2004),
Hidayat and Rehmayani (2007) reported that optimal transmission of TLCV, PepYLCIV, and ToLCV required at least 10 individual whiteflies per plant with 24 hr acquisition and 48 hr inoculation feeding period. Transmission of
Begomovirus by mechanical inoculation or through dodder plant and seed was not
proven (Nariani 1960; Bedford et al. 1994) although Salam (2005) reported
8
B. tabaci is a polyphagous insect pest, has a wide host range, and an important vector of virus species belonging to Begomovirus (90%), Crinivirus
(6%), Closterovirus and Carlavirus (4%) (Jones et al. 2003). Sulandari et al.
(2006) reported infection of PepYLCIV using whitefly transmission on plant species from the families Solanaceae, Compositae, and Leguminosae. Cucurbit, chili pepper, tomato, and weed species Euphorbia heterophylla are potential for
whitefly reservoir as well as Begomovirus host in Nicaragua (Rojas 2004).
Another important Begomovirus, Cotton leaf curl virus, can be transmitted to several plants, i.e. cotton (Gossypium hirsutum), tomato (Lycopersicon esculentum), Ageratum conyzoides, and Sida alba (Kang et al. 2004). Legume
infecting Begomovirus, MYMV and MYMIV, were able to infect some Leguminosae, such as mungbean (Vigna radiata), blackgram (Vigna mungo), pigeonpea (Cajanus cajan), soybean (Glycine max), mothbean (Vigna aconitifolia), and common bean (Phaseolus vulgaris). Single infection of MYMV and MYMIV was found in yard long bean (Vigna unguiculata subsp.
sesquipedalis) and Lablab purpureus, respectively (Qazi et al. 2007). MYMIV
was also observed infecting two wild species of Vigna, i.e. V. hainiana and V. trilobata (Naimuddin et al. 2011).
Organization and Molecular Characteristics of Begomovirus
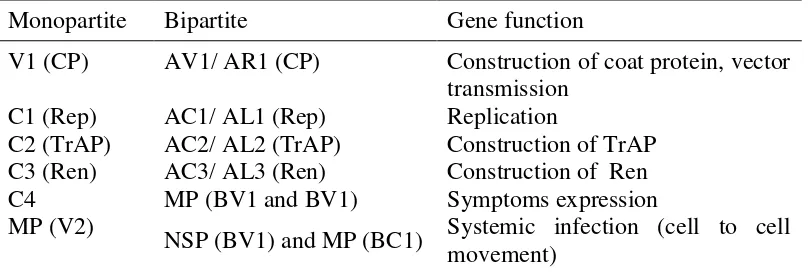
Begomovirus genome have six to seven open reading frames which encode proteins with specific function (Table 1, Fig 2). Bipartite genome of Begomovirus
is referred to DNA-A and DNA-B. DNA-A is required for DNA replication and encapsidation whereas DNA-B encodes systemic infection and symptoms expression. The combination of DNA-A and DNA-B open reading frame is present in monopartite genome with similar function as in bipartite genome (Hull 2002; Fauquet et al. 2005; Rojas et al. 2005).
Table 1 Genome organization of Begomovirus (Hull 2002)a
Monopartite Bipartite Gene function
V1 (CP) AV1/ AR1 (CP) Construction of coat protein, vector transmission
C1 (Rep) AC1/ AL1 (Rep) Replication
C2 (TrAP) AC2/ AL2 (TrAP) Construction of TrAP C3 (Ren) AC3/ AL3 (Ren) Construction of Ren
C4 MP (BV1 and BV1) Symptoms expression
MP (V2) NSP (BV1) and MP (BC1) Systemic infection (cell to cell movement)
a
CP: coat protein, Rep: replication-associated protein, TrAP: transactivating protein, Ren: replication enhancer.
9 encodes Rep, an initiator for rolling circle replication (Lazarowitz 1987; Harrison and Robinson 1999; Usharani et al. 2004). The nonanucleotide is conserved,
always found on loop sequence in the “stem-loop structure”. The “stem-loop
structure” was formed at nucleotide number 29-32, GGCA(T/A)CCGN(T/A)(A/T)TAATATTACCGG(A/T)TGGCC which was almost found in dicotyledonous host. This sequence motif regulates the compatibility between virus and host cell and gene expression (Lazarowitz 1987).
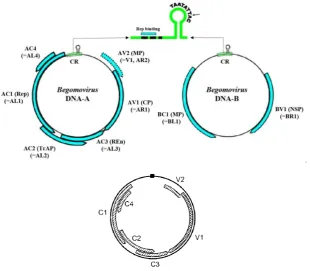
Fig 2 Genome organization of Begomovirus: bipartite (Cuong 2007) (above) and monopartite (Hull 2002) (below)
CR of Begomovirus also contains repetitive sequence and TATA box
motif. Repetitive sequence, called as “Iteron”, commonly has AC/TC/TGGTGT
motif which works high specifically with nonanucleotide in Rep binding site (Argüelo-Astorga et al. 1994; Chatterji et al. 2000; Yadava et al. 2010). Bean infecting-Begomovirus and almost old world Begomoviruses has one iteron with
an invert repeat (downstream iteron) and two tandem iteron before TATA box motif (upstream iterons). The downstream iteron is absent in the new world
Begomovirus (Argüelo-Astorga et al. 1994; Usharani et al. 2004). Diagnosis and Detection of Begomovirus
Begomovirus has been described as non-seed transmitted, only transmitted
10
microscope (Boss 1990). Detection of Begomovirus is commonly done using molecular method.
ELISA (Enzyme – Linked Immunosorbent Assay) is one of sensitive and
common serological methods used for plant virus detection (Clark and Adams 1977). Detection of Begomoviruses using ELISA was reported for ACMV, EACMV (Thomas et al. 1986; Harrison et al. 2002), BGMV (Harrison et al.
2002; Cancino et al. 1995), BDMV, TYLCV (Harrison et al. 2002), ToLCV,
ToLCBV (Devaraja et al. 2003), PepYLCIV (Sulandari 2004). Another serological method, Dot Immunobinding Assay (DIBA), has also been reported to detect TYLCV (Lapidot 2002; Kushwaha et al. 2010) and PepYLCIV (Hidayat et al. 2009). Cross reaction and low specificity are the reasons why serological method is not preferred for Begomovirus detection (Kushwaha et al. 2010).
More common method to detect Begomovirus infection is based on Polymerase Chain Reaction (PCR) technique using degenerate primers. Several degenerate primers such as PAL1v1978/ PAR1c715 (Rojas et al. 1993), Deng A/ Deng B (Deng et al. 1994), AV494/ AC1048 (Wyatt and Brown 1996), SPG1/
SPG2 (Li et al. 2004) have been successfully used for variants of Begomovirus.
PCR based detection of Begomovirus using degenerate primer has also reported in Indonesia. The above degenerate primers have been used to identify and characterize various members of Begomovirus, i.e. PePYLCIV from chili pepper
(Hidayat et al. 1999; Sulandari et al. 2006; Trisno et al. 2009), TLCV from tobacco (Hidayat et al. 2008), TYLCV from tomato (Aidawati et al. 2005).
Amplification of Begomovirus on the top region using PCR is important to
identify character of Begomovirus based on sequence analysis. Fragment of Begomovirus DNA amplified by degenerate primer PAL1v1978/ PAR1c715 covers part of AL1 region (replicate gene), common region and intergenic region, and part of AR1 region (coat protein gene) (Rojas et al. 1993). Nucleotide sequence can be used for identification based on CR which has been described before as identity character of every Begomovirus species (Lazarowitz 1987;
Rojas et al. 1993). Study of CR in different Begomovirus species showed distinct
character, i.e. iteron sequence, position number of TATA box sequence, and hair pin loop structure. Characterization of Begomovirus based on CR in Indonesia has
been conducted by Hidayat et al. (2008) for TLCV and Trisno et al. (2009) for
PepYLCIV. TLCV has different CR character with PepYLCIV, especially in hairpin loop structure (Fig 3).
Fig 3 Hair pin loop structure of Begomovirus: A and B, PepYLCIV (Trisno et al.
2009); C, TLCV (Hidayat et al. 2008)
B C
11
MATERIALS AND METHODS
Field Survey and Samples Collection
Surveys were conducted in West Java (Bogor and Subang), Central Java (Tegal, Klaten, and Magelang), and Jogyakarta (Sleman and Bantul) on March 2012. Leaf samples were collected using purposive sampling method based on typical symptoms as described by Damayanti et al. (2009) and Ilyas et al. (2010),
i.e. leaf yellowing and yellow mosaic. Fresh tissue was directly subjected for virus detection and the remainings were stored at -80oC as isolate collection in the laboratory.
Detection and Identification of Viruses from Infected Leaves
Detection of viruses from infected leaves was conducted using Indirect enzyme linked-immunosorbent assay (I-ELISA) and polymerase chain reaction (PCR) method. The objective was to determine the current status of Begomovirus
and other viruses associated with yellow mosaic disease in yard long bean. Virus Detection by I-ELISA
Two major viruses on yard long bean, i.e. Potyvirus and CMV, was detected
from leaf samples using I-ELISA commercial kit following manufacturer protocol (Leibniz-Institut DSMZ GmbH, Germany).
Infected leaves was ground in coating buffer (1/10, v/v). Coating buffer (pH 9.6) contains of 15 mM Na2CO3, 2.38 mM NaHCO3, 3.08 mM NaN3, and H2O was added to final volume of 1 L. Aliquots of each sample (100 µl) was dispensed
12
Begomovirus Detection by PCR
Extraction of Total Viral DNA. Total viral DNA was isolated from infected leaf following a procedure described by Doyle and Doyle (1987) with minor modification. Fresh tissue (0.1 g) was ground in liquid Nitrogen to powder, 500 µl of CTAB buffer (10% Cetyl-trimethyl-ammonium bromide, 0.1 M Tris-HCl pH 8, 0.05 M EDTA, 0.5 M NaCl, 1% β-mercapto-ethanol) was added, and the sap was transferred to 1.5 ml clean tube. The sap was incubated in water bath at 65 oC for 1 hr, then shaked every 10 min to separate lipid and protein. 500 µl of chloroform/ iso-amyl alcohol (24:1, v/v) was added to the liquid, then tube was vortexed for 5 min, and centrifuged at 14 000 rpm for 15 min. The supernatant was pipetted to 1.5 ml clean tube, 3 M ammonium acetate and isopropanol of 1/10 and 2/3 volume supernatant, was added respectively. The liquid was mixed gently then incubated overnight at -20 oC or 4 hr at room temperature. After incubation, the liquid was centrifuged at 12 000 rpm for 10 min to precipitate DNA and then discarded flow-through. The pellets were washed with 500 µl of 70% ethanol, centrifuged at 8000 rpm for 5 min and dried under room temperature after discarding the flow through. Dried pellets containing total DNA were dissolved in 50 to 100 µl of nuclease free water or TE buffer (pH 8) and the DNA was ready for amplification.
Viral DNA Amplification. Amplification of viral DNA-A and DNA-B was
conducted following method described by Rojas et al. (1993) to confirm
Begomovirus infection (Table 2). PCR reaction contains 10 x PCR Buffer, 25 mM MgCl, 2.5 mM dNTPS, 10 µM each of primer, Taq polymerase (5 U/µl), 1 µl of
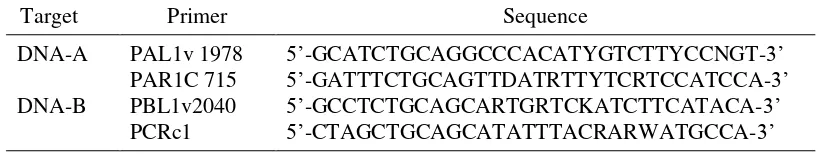
DNA, and the reaction was adjusted to 25 µl with nuclease free water. Amplifications was performed in GeneAmp PCR System 9700 machine with 5 min at 94.0 oC for pre-heating, followed by 30 cycles of denaturation (1 min at 94.0 oC), annealing (1 min at 50.0 oC), and extension (3 min at 72.0 oC). The last cycle was ended at 72.0 oC for 3 min and cooled down to 4.0 oC. Electrophoresis was done using 1% Agarose gel in 0.5 x TBE (Tris-Boric acid-EDTA) buffer, run at 50 V for 50 min. Following electrophoresis, agarose gel then was soaked on to 0.1% EtBr for 5 min, washed with H2O, and visualized under UV transilluminator. Table 2 Degenerate primers for amplification of Begomovirus using polymerase
chain reaction (Rojas et al. 1993)
Target Primer Sequence
DNA-A PAL1v 1978 5‟-GCATCTGCAGGCCCACATYGTCTTYCCNGT-3‟
PAR1C 715 5‟-GATTTCTGCAGTTDATRTTYTCRTCCATCCA-3‟
DNA-B PBL1v2040 5‟-GCCTCTGCAGCARTGRTCKATCTTCATACA-3‟
PCRc1 5‟-CTAGCTGCAGCATATTTACRARWATGCCA-3‟
Cloning Strategy of DNA-A of Begomovirus
DNA Purification. Partial DNA-A fragments obtained from amplification above will be used for cloning. Sample was selected based on quality of amplification product. Selected sample from Tegal (pTgl) was purified according
13 rpm for 1 min, discarded flow – through and replaced the column to tube collection. PE buffer (30 µl) was added and column was centrifuged at 14 000 rpm for 1 min, discarded flow – through and replace the column to tube collection. Column was centrifuged again at 14 000 rpm for 1 min, column was placed to 1.5 ml clean tube and then added 30 µl EB buffer to elute DNA. Column was incubated for 1 min and then centrifuged at 14 000 rpm for 1 min to get pure DNA.
Ligation. Pure DNA was mixed with pCR®4-TOPO® vector according to
TOPO TA Cloning Kit for Sequencing‟s instructions (InvitrogenTM – Life Technologies Corp., Carlsbad – California, US). On to pure DNA (0.5 – 4 µl) was
added 1 µl salt solution and water up to 5 µl (water didn‟t need if in the reaction
used 4 µl pure DNA). The reagent was mixed by pipetting up and down, then added 1 µl pCR®4-TOPO® vector. The reagent was incubated at 23 – 25 oC for 30 min to get plasmid vector for transformation.
Fig 4 pCR®4-TOPO® vector for cloning (InvitrogenTM – Life Technologies Corp., Carlsbad – California, US)
Transformation. Transformation was conducted to insert plasmid vector into competent cell using One Shot TOP 10 and DH5α-TIR Competent Cells. Plasmid vector (2 µl) was mixed gently with competent cell and incubated at 42 oC for 30 sec then immediately transferred to ice (heat shock culture method). On to competent cell was added 250 µl SOC medium (triptone, yeast extract, NaCl, glucose, 250 mM KCl, H2O), shaked and the mixture was incubated at 37 oC for 60 min.
14
incubated overnight at 37 oC. Selected colony from media was incubated on LB medium which contains ampicillin (1 µl of 100 mg/ml ampicillin stock per 2 ml LB). LB medium was shaked at 37 oC overnight to grow the culture to log phase.
Plasmid DNA Isolation. Plasmid DNA was isolated using two methods, i.e. colony PCR and miniprep culture. Colony PCR was conducted by touching colony with pipette tip then resuspended in 100 µl H2O to new tube, and use 1 µl suspension as template in PCR reaction. Miniprep culture was conducted using QIAprep Spin Miniprep Kit (Qiagen Inc., Valencia – California, US). Pellet 1.5 ml bacterial overnight culture was centrifuged at 14 000 rpm for 3 min at room temperature (15-25 oC) and then discarded the liquid flow-through. Pelleted bacteria was resuspended in 250 µl buffer P1 (contains LyseBlue® reagent) and transferred to a micro-centrifuge tube. Buffer P2 (250 µl) was added to the tube and mixed thoroughly by inverting the tube 4 – 6 times until the solution turned blue (do not allow the lysis reaction to proceed for more than 5 min). Buffer N3 (350 µl) was added and mixed immediately and thoroughly by inverting 4 – 6 times until the solution turned colorless. The tube was centrifuged for 10 min at 13 000 rpm. The supernatant was transferred to QIAprep spin column by pipetting then centrifuged at 14 000 rpm for 1 min. The liquid was discarded flow through, added 750 µl buffer PE, centrifuged at 14 000 for 1 min, then discarded the liquid flow through. The column was replaced to the new 1.5 ml tube and added 30 µl buffer EB to elute DNA, let the tube stand for 1 min, centrifuged at 14 000 rpm for 1 min to get plasmid DNA. Plasmid DNA was digested using EcoR I enzyme. Digestion reaction contains 0.2 µl EcoR I, 2 µl buffer 4, plasmid DNA, and H2O per reaction. Isolated clones from PCR colony and miniprep culture were visualized using electrophoresis gel as described earlier.
DNA Sequencing
Viral DNA fragments obtained from direct PCR amplification and plasmid DNA isolation were sent to PT. Genetika Science, Indonesia and Australian Genome Research Facility, Australia, respectively for DNA sequencing. Sequence data were compared with other sequences from GenBank (NCBI 2013) and analyzed using software programs BioEdit V.7.0.5, CLC Sequence Viewer 7, and MEGA 6.06.
Construction of Specific Primer
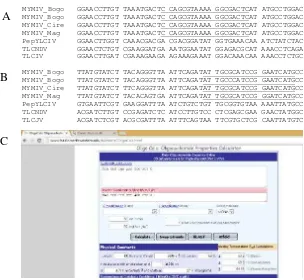
Primers were constructed by two methods, (1) semi-manual OligoCalc: Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/ biotools/OligoCalc.html) and (2) primer BLAST program on NCBI (NCBI 2013). The first method was conducted by aligning sequences of Begomovirus infecting yard long bean and other Begomoviruses from Indonesia reported earlier using BioEdit, followed by selecting conserved region for only Begomovirus infecting
yard long bean. The selected nucleotides were analysed on OligoCalc for their property. Forward (positive sense) and reverse (negative sense) primers were determined as MYF and MYR, respectively (Table 3 and Fig 5).
15 sense) primers were determined as MY1 and MY2, respectively (Table 3 and Fig 6).
Table 3 Specific primers for amplification of Begomovirus infecting yard long bean
Code Sequence (5‟ to 3‟) Position at Genome
of Begomovirus
Expected Product (bp) MYF
MYR
F:CCAGCGTAAAAGGCGACTC
R:CATGATTCCGGATGCGCAAT
2068 – 2086
318 – 299 ± 1000 bp MY1
MY2
F:TTACATGGTCCCTCGCAACC
R:ACAGCCTTCTCTACCCCGAT
195 – 214
395 – 375 ± 238 bp
MYMIV_Bogo GGAACCTTGT TAAATGACTC CAGCGTAAAA GGCGACTCAT ATGCCTGGAC
MYMIV_Bogo GGAACCTTGT TAAATGACTC CAGCGTAAAA GGCGACTCAT ATGCCTGGAC MYMIV_Cire GGAACCTTGT TAAATGACTC CAGCGTAAAA GGCGACTCAT ATGCCTGGAC MYMIV_Mag GGAACCTTGT TAAATGACTC CAGCGTAAAA GGCGACTCAT ATGCCTGGAC PepYLCIV GGAACTTGGT CAAACGACGA CGACGGATAT GGTGAAACAA ATCTATCTAC TLCNDV GGAACTCTGT CGAAGGATGA AATGGAATAT GGAGACGCAT AAACCTCAGA TLCIV GGAACTTGAT CGAAAGAAGA AGAAAGAAAT GGACAAACAA AAACCTCTGC
MYMIV_Bogo TTATGTATCT TACAGGGTTA ATTCAGATAT TGCGCATCCG GAATCATGCC MYMIV_Bogo TTATGTATCT TACAGGGTTA ATTCAGATAT TGCCCATCCG GAATCATGCC MYMIV_Cire TTATGTATCT TTCAGGGTTA ATTCAGATAT TGCGCATCCG GAATCATGCC MYMIV_Mag TTATGTATCT TACACAGTGA ATTCAGATAT TGCGCATCCG GGATCATGCC PepYLCIV GTGAATTCGT GAAGGATTTA ATCTGTCTGT TGCGGTGTAA AAATTATGCC TLCNDV ACGATCTTGT CCGAGATCTC ATCCTTGTCC CTCGAGCGAA GAACTATGGC TLCJV ACGATCTCGT ACGCGATTTA ATTTCAGTAA TTCGTGCTCG CAATTATGTC
Fig 5 Alignment of conserved region to design specific primer for Begomovirus
infecting yard long beans using BioEdit V.7.0.5. A, sequence of forward primer (MYF) indicated by underlined letters; B, sequence of reverse primer indicate underlined letters before swap strand (MYR) using OligoCalc; C, primer analysis using OligoCalc.
A
B
16
Fig 6 Construction of specific primer using primer BLAST program at NCBI Evaluation of Primer Specificity
Primer pairs which were designed in this research, MYF/ MYR and MY1/ MY2 were evaluated for their capability to amplify specific target, i.e.
Begomovirus infecting yard long bean. Amplification was conducted as described earlier with modification on annealing condition, i.e. 59.5 oC for MYF/ MYR and 61 oC for MY1/ MY2. Three type of samples was included in this evaluation: (1)
Begomovirus from yard long bean, (2) Begomovirus samples infecting other crops in Indonesia (PepYLCIV from chili pepper, ToLCNDV from tomato, and ToLCKV from eggplant), (3) non Begomovirus samples (RTBV from rice field
and BCMV which infected yard long bean). Amplification was undergone following method that has been described before (Rojas et al. 1993) with
annealing temperature of 59.5 oC for MYF/ MYR and 61 oC for MY1/ MY2.
Koch’s Postulate
Koch‟s postulate was conducted to proof Begomovirus as the causal agent of
yellow mosaic diseases on yard long bean. Infected yard long bean from the field was used as source of virus inoculum for transmission using whitefly (B. tabaci). Whitefly transmission involved acquisiton feeding period (isolation) by placing the whitefly on infected plants for 24 hr, followed by inoculation feeding period on healthy plants for 48 hr by infesting 10 viruliferous whitefly to each plant.
Three varieties of yard long bean, i.e. „Parade‟, „New Jaliteng‟, and „Wulung‟
were used for this study. Confirmation of Begomovirus infection was conducted
by PCR-based detection method using specific primer MY1/MY2 two weeks after inoculation.
This transmission study showed the principle of Koch‟s Postulate, i.e. (1)
The microorganism must be found in abundance in all organisms suffering from the disease, but not be found in healthy organisms; (2) The microorganism must be isolated from a diseased organism and grown in a pure culture; (3) The cultured microorganism should cause disease when introduced into a healthy organism; (4) The microorganism must be reisolated from the inoculated, diseased host and identified as being identical to the original microbe (Koch 1876)
17
RESULT AND DISCUSSION
Identification of Begomovirus from Field Samples
Incidence of yellow mosaic disease was very high, i.e. 80% to 100% in most growing areas. Infected plants were easily recognized in the field based on visual symptoms. Four main symptoms of yellow mosaic disease in the field were observed, i.e. (1) yellowing, (2) yellowing with green spot, (3) yellow vein netting, and (4) mosaic vein banding (Fig 7). The most common symptom found in every field was yellowing. Yellowing with green spot was thought as early symptom before it develops to become yellowing. Further severe infection caused smaller pods and leaves. Yellow vein netting was only found in Tegal in one plant and therefore the sample was not used for further detection.
Fig 7 Various symptoms found associated with yellow mosaic disease on yard long bean : yellowing (A), yellowing with green spot (B), yellow vein netting (C), mosaic vein banding (D)
Begomovirus was detected in 11 infected plant samples (Fig 8) showing
yellowing and yellowing with green spot symptoms, 4 of them has mixed infection with Potyvirus (Table 4). Two samples from Subang showing mosaic vein banding symptoms was infected only by Potyvirus. Mosaic vein banding
symptom caused by Potyvirus infection has been described previously by
Damayanti et al. (2009) and Melinda (2013). Infection of CMV was not detected from all samples. This result indicated Begomovirus as the major virus associated with yellow mosaic disease of yard long bean in Java.
C D
18
DNA fragment of ± 1.6 kb was successfully amplified using PAL1v1978/ PAR1c715 from 11 out of 14 samples (Fig 8). Intensity of DNA bands on the agarose gel was varied, i.e. strong (samples from Tegal and Bogor), medium (samples from Magelang), and weak (samples from Klaten, Sleman, and Subang). Due to its quality, DNA fragment origin of samples from Tegal was selected for cloning.
Fig 8 Visualization of PCR products of Begomovirus amplification from leaf samples using universal primers PAL1v1978/ PAR1c715 on 1 % agarose gel. M, 1 kb DNA marker (Thermo Scientific, US)
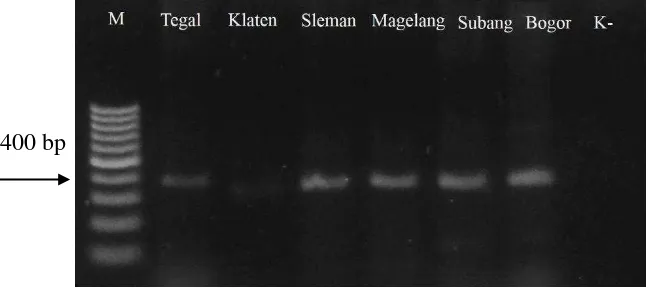
DNA-B fragment of ±400 bp was successfully amplified from 6 samples, i.e. from Tegal, Klaten, Sleman, Magelang, Subang, and Bogor using degenerate primer for DNA B, PBL1v2040/PCR1c (Fig 9). The size of DNA-B fragment was different with those reported previously by Rojas et al. (1993), however it was
still acceptable as explained by Rojas et al. (1993). Strong bands of PCR product
was shown on amplification using samples from Tegal, Sleman, Magelang, Subang, and Bogor, whereas amplification using sample from Klaten resulted weak band of PCR product. Amplification of DNA-B proved the bipartite nature of Begomovirus infecting yard long bean in Java.
Fig 9 Visualization of PCR products of Begomovirus amplification from leaf
samples using universal primers PBL1v2040/ PCR1c on 1 % agarose gel. M, 100 bp DNA marker (Thermo Scientific, US)
± 1.6 kb
19 Table 4 Detection of CMV, Potyvirus, and Begomovirus from leaf samples
collected from various locations using I-ELISA and PCRa
a -, not detected; +, detected Cloning of DNA A Begomovirus
Isolate from Tegal was selected for DNA cloning based on its quality of amplified DNA, i.e. best DNA concentration and discrete DNA band (Fig 4). Three DNA clones were obtained, determined as pTgl1, pTgl2 and pTgl3. Confirmation of insert DNA was done using colony PCR and restriction endonuclease digestions of plasmid preparation (Maniatis et al. 1982). Colony
PCR using primer M13F/M13R and plasmid digestion using EcoR I enzyme confirmed the insert DNA (Fig 10). One DNA clone (pTgl1) was selected for further nucleotide sequence analysis and construction of specific primer.
Cloning using plasmid pCR®4-TOPO® is preferable because it allows direct selection of recombinants via disruption of the lethal E. coli gene, ccdB. Ligation of a PCR product disrupts expression of the lacZα-ccdB gene fusion
permitting growth of only positive recombinants upon transformation in TOP10 cells. Cells that contain non-recombinant vector are killed upon plating (Invitrogen 2006).
Location Symptoms description I-ELISA PCR
CMV Potyvirus Begomovirus
Tegal Yellowing - + +
Yellowing mosaic - - +
Klaten Yellowing - - +
Yellowing mosaic - - -
Sleman Yellowing - + +
Yellowing mosaic - + +
Magelang Yellowing mosaic - - +
Yellowing - + +
Yellowing mosaic - - +
Subang 1 Mosaic vein banding - + -
Mosaic vein banding - + -
Subang 2 Mosaic vein banding - - -
Yellowing - - +
Bogor Yellowing - - +
20
A B
Fig 10 Visualization of PCR product from Begomovirus clone (pTgl) using colony PCR (A); digestion of pTgl clone using EcoR I enzyme (B); M,
marker 1kb plus
Sequence Identity Analysis of Begomovirus Infecting Yard Long Bean
Nucleotide sequences were obtained for Begomovirus isolates from Tegal,
Klaten, Magelang, Subang, Cirebon, and Bogor. Analysis of their identity by comparing to sequences on the GenBank showed their highest homology with
Mungbean yellow mosaic India virus (MYMIV) from Brebes and Purwakarta, i.e.
>95%, followed by MYMIV from Bangladesh, Nepal, Pakistan, and India, i.e. >88% (Table 5). Their homology to MYMV, another virus causing yellow mosaic disease in South Asia, is only 73% - 77% and to other Begomovirus
reported from Indonesia is even lower, i.e. 53% - 57%.
Table 5 Nucleotide sequence homology (%) of Begomovirus infecting yard long bean in Java with other Begomoviruses reported earlier in GenBank
Begomovirus infecting yard long bean
Begomovirus isolates from GenBank*)
1 2 3 4 5 6 7 8 9 10 11 12
pTgl1 99.7 99.8 93.0 94.4 94.2 94.1 77.1 57.8 55.4 53.6 56.6 19.0 Tegal 2 95.7 95.8 89.3 90.7 90.6 90.5 74.4 55.5 54.7 53.7 56.3 19.6 Klaten 96.0 96.0 89.5 91.0 90.9 90.8 74.5 55.8 55.6 54.5 57.0 19.3 Magelang 95.9 95.9 89.3 90.9 90.7 90.7 74.4 55.1 55.4 54.2 56.6 19.1 Magelang 2 95.3 95.3 88.8 90.3 90.1 90.0 74.1 54.6 54.8 53.6 56.4 19.5 Subang 95.5 95.7 89.2 90.6 90.4 90.4 74.2 55.4 54.7 53.7 56.3 19.6 Bogor 1 95.4 95.4 89.1 90.3 90.2 90.1 74.2 55.0 55.1 54.0 56.7 19.6 Bogor 2 95.1 95.3 88.8 90.2 90.0 90.0 73.8 55.2 54.5 53.8 56.1 19.7 Cirebon 95.7 95.8 89.3 90.7 90.6 90.5 74.2 55.6 54.9 53.8 56.5 19.6
*)1: MYMIV Brebes (JN368436); 2: MYMIV Purwakarta (JN368434); 3: MYMIV India
(KC852204); 3: MYMIV Bangladesh (AF314145); 5: MYMIV Nepal (AY271895); 6: MYMIV Pakistan (AM992618); 7: MYMV India (KC911271); 8: PepYLCIV (AB246170); 9: TLCIV (AB241671), 10: ToLCNDV (data unpublished); 11: ToLCJV (189848); 12: BCMV Bogor (FJ653916)
pTgl K- M
pTgl undigest M