Resistance to bacterial wilt in somatic hybrids between
Solanum
tuberosum
and
Solanum phureja
I. Fock
a, C. Collonnier
a, A. Purwito
b, J. Luisetti
c, V. Souvannavong
d, F. Vedel
e,
A. Servaes
a, A. Ambroise
a, H. Kodja
f, G. Ducreux
a, D. Sihachakr
a,*
aMorphogene`se Ve´ge´tale Expe´rimentale,Baˆt.360Uni6ersite´ Paris Sud,91405Orsay Cedex,France
bPlant Biomolecular and Cellular Laboratory,IUC Biotechnology,Bogor Agricultural Uni6ersity(IPB),JL Puspa Kampus IPB Darmaga, Bogor16610,Indonesia
cLaboratoire de Phytopathologie,7chemin de l’IRAT,Ligne Paradis,97410Saint-Pierre,La Re´union,France dGroupe Endotoxines,UMR8619,CNRS-UPS,Baˆt.430Uni6ersite´ Paris Sud,91405Orsay Cedex,France
eLaboratoire d’Ecologie,Syste´matique et E6olution,UPRESA-CNRS8079,Baˆt.360Uni6ersite´ Paris Sud,91405Orsay Cedex,France fLaboratoire de Biologie et Physiologie Ve´ge´tales,Ge´ne´tique Mole´culaire et E6oluti6e,15A6enue Rene´ Cassin,BP7151,97715Saint-Denis,
La Re´union,France
Received 2 March 2000; received in revised form 25 August 2000; accepted 4 September 2000
Abstract
Somatic hybrid plants were produced after protoplast electrofusion between a dihaploid potato, cv. BF15, and a wild tuber-bearing relative, Solanum phureja, with a view to transferring bacterial wilt resistance into potato lines. A total of ten putative hybrids were selected. DNA analysis using flow cytometry revealed that six were tetraploids, two mixoploids, one amphiploid and one octoploid. In the greenhouse, the putative hybrids exhibited strong vigor and were morphologically intermediate, including leaf form, flowers and tuber characteristics. The hybrid nature of the ten selected plants was confirmed by examining isoenzyme patterns for esterases and peroxidases, and analysis of RAPD and SSR markers. Analysis of chloroplast genome revealed that eight hybrids possessed chloroplast (ct) DNA of the wild species, S. phureja, and only two contained
Solanum tuberosum ct type. Six hybrid clones, including five tetraploids and one amphiploid, were evaluated for resistance to bacterial wilt by using race 1 and race 3 strains ofRalstonia solanacearum, originating from Reunion Island. Inoculations were performed by an in vitro root dipping method. The cultivated potato was susceptible to both bacterial strains tested. All somatic hybrids except two were tolerant to race 1 strain, and susceptible to race 3 strain. Interestingly, the amphiploid hybrid clone showed a good tolerance to both strains. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Solanum tuberosum;Solanum phureja; Electrofusion; Somatic hybrids; Bacterial wilt;Ralstonia solanacearum
www.elsevier.com/locate/plantsci
1. Introduction
Bacterial wilt, a severe and devastating plant disease caused by Ralstonia solanacearum, occurs in most tropical, subtropical and warm temperate areas and even in some cool temperate regions [1]. It affects more than 200 plant species distributed in more than 50 families, particularly potatoes and tomatoes [2].
The bacterium invades plant vascular tissues from wounded roots or natural openings (sec-ondary roots). The colonisation of the stem results in browning of the xylem and frequently leads to partial or complete wilting [3]. Potatoes and toma-toes are affected by two of the three major races of
R. solanacearum. Under cool temperate condi-tions, particularly in north-western Europe or at high altitude in some tropical countries such as Peru or Reunion Island, only race 3 strains induce wilting of potatoes [4]. Race 1 strains also damage potato crops under tropical and subtropical condi-tions [5].
* Corresponding author. Tel.:+33-1-6915-4690; fax:+ 33-1-6985-5490.
E-mail address:[email protected] (D. Sihachakr).
Resistant or tolerant cultivars are mainly uti-lized for disease management since chemicals are not effective and sanitation measures difficult to apply. Therefore, the worldwide control strategy has consisted of plant breeding for resistance to bacterial wilt [6]. Using resistant cultivars to con-trol bacterial wilt has been successful for tobacco and peanut. Since immunity was not identified in potato, only tolerant cultivars were selected [7], such as cultivar Prisca mainly cropped in Mada-gascar, cultivar Ndinamagara in Burundi, Rwanda and Zaire and cultivar Achat in Brazil [4,8]. To better control bacterial wilt, continuous develop-ment of resistant or tolerant varieties is needed. Some wild or related cultivated species are known to be resistant or highly tolerant to bacte-rial wilt and thus are potential sources for resis-tance. Unfortunately, hybrids of potato with resistant genotypes of Solanum chacoense,
Solanum sparsipillum and Solanum multidissectum
revealed some traits of wildness such as a high glycoalkaloid content besides a moderate level of resistance to bacterial wilt [4]. Unlike these wild species which have been classified as wildTuberosa
[9],Solanum phureja (cultivated Tuberosa) is phyl-logenetically close to Solanum tuberosum and dis-plays resistant traits, dominant and readily transmitted to progeny. Some clones of S. phureja
with high degree of resistance to bacterial wilt have been used for crossing with commercial culti-vars ofS. tuberosum[4,10]. Resistance to bacterial wilt derived from S. phureja was first described as dominant and controlled by three unlinked genes [6,11,12]. More recently, at least four major genes have been reported to be involved in potato resis-tance to bacterial wilt [13].
The introgression of multiple resistance genes from wild Solanum species into S. tuberosum by classical breeding methods is time-consuming, la-borious and may encounter difficulties because of sexual incompatibilities, particularly due to differ-ences in ploidy level or in endosperm balance numbers. Somatic fusion is expected to provide a new possibility for increasing genetic variability, and also a means of transferring desirable agro-nomic traits into potato. The potential use of somatic hybridisation has been demonstrated by the successful introduction of traits such as resis-tance to viruses [14,15] and frost [16] from
Solanum bre6idens, resistance toPhytophtora infes
-tans and Globodera pallida from Solanum circaei
-folium [17], insect resistance from Solanum berthaultii [18], and resistance to bacterial wilt from Solanum commersonii[19], into the cultivated potatoes. In other studies of fusion between potato and S. phureja, partial elimination of chro-mosomes was reported in the resulting somatic hybrids. The nuclear chromosomes of S. phureja
were preferentially eliminated [20,21]. So far, no information has been available about evaluation of the somatic hybrid clones for the introgression of resistance against bacterial wilt from S. phureja
into potato.
In this study, somatic hybridisation between the dihaploid S. tuberosum cv. BF15 and the diploid
S. phureja was performed. The somatic hybrids were identified and characterised as well as evalu-ated for resistance to race 1 and race 3 strains of
R. solanacearum.
2. Materials and methods
2.1. Plant material
A clone of dihaploidS. tuberosum L. (2n=2× =24 chromosomes), cv. BF15, obtained from the Institut National de a Recherche Agronomique (INRA), Landerneau, France, and a diploid clone of the wild tuber-bearing S. phureja, SVP5(PH77-1445-2242), provided by the Centre for Genetic Resources, Wageningen, the Netherlands, were used. Plants were maintained in vitro from axillary shoot tips on MS basal medium [22] as described by Chaput et al. [23]. The clones were subcultured at 4-week intervals. Environmental conditions were 14 h day−1 illumination at 55
mmol m−2
s−1, 20°C and 60% relative humidity.
2.2. Protoplast isolation and electrofusion
resulting suspension was purified and rinsed by centrifugation in successive 0.6 M sucrose, and 0.5 M mannitol+0.5 mM CaCl2 solutions. Prior to electrical fusion, the protoplasts were suspended in the last washing solution, and the density for both species adjusted to 4.0×105 protoplasts ml−1.
Electrical apparatus and fusion procedure were described previously [25]. The movable multi-elec-trodes were placed in a 15×50-mm Petri dish containing 500 – 700ml of a mixture (1:1) of
proto-plasts from both parents. The protoproto-plasts were aligned for 15 s by the application of an a.c. field at 230 V cm−1 and 1 MHz. Subsequently, one square pulse developing 1.2 kV cm−1 for 40
ms
was applied to achieve protoplast fusion. After application of the square pulse, the a.c. field was reduced routinely to 20 V cm−1to keep alignment of protoplast chains and to estimate fusion fre-quency by observing fusion events through an inverted microscope.
2.3. Protoplast culture and plant regeneration
The culture medium was VKM medium [26] supplemented with 250 mg l−1 PEG, 0.2 mg l−1 2,4-D, 0.5 mg l−1 zeatin, 1 mg l−1 NAA, 0.2 M mannitol and 0.2 M glucose as osmotic agents and 0.05% (w/v) MES. The pH was adjusted to 5.8 prior to sterilizing by filtration (0.22-mm filter,
Millipore). After electrical treatment, 6 ml of cul-ture medium were added progressively to the Petri dish containing the fused protoplast mixture. Cul-tures were kept in the dark for 7 days prior to putting them into the light. On day 15, cultures were diluted eight times with fresh VKM medium supplemented with 2 mg l−1BAP and 0.1 mg l−1 2,4-D, pH 5.8. Calli (3 – 4 mm diameter) were then transferred onto the regeneration medium: MS basal medium, plus vitamins [27], to which was added 2% (w/v) sucrose, 2 mg l−1 zeatin and 0.1 mg l−1 IAA and solidified with 7 g l−1 agar (Difco). Emerging shoots were excised from callus and plantlets were multiplied by subculturing leafy node cuttings on hormone-free MS medium. Both parental and selected hybrid plants were trans-ferred to the greenhouse. Environmental condi-tions were 14 h day−1 illumination at 55
mmol
m−2 s−1, 20°C and 60% relative humidity for in vitro cultures and 16 h day−1 illumination at 180 mmol m−2 s−1, 25 – 30°C and 70 – 85% relative
humidity in the greenhouse.
The selection and identification of somatic hy-brids was based on the analysis of plant morphol-ogy and ploidy level. Evidence for the hybrid nature of selected plants was provided by examin-ing isoenzyme patterns and DNA analysis.
2.4. Determination of ploidy le6el and pollen 6iability
Flow cytometry was used to quantify DNA for the determination of ploidy level. Using a razor blade 1 cm2of leaf material from in vitro plants was chopped in 1 ml buffer containing CPW salts [24], 0.5 M mannitol, 0.25% (w/v) PEG, 0.5% (w/v) Triton X-100, 0.25% (v/v) mercaptoethanol at pH 6.5 – 7. Crude samples were filtered through a 40-mm mesh nylon and stained with 4,6
di-amidino-2-phenylindole (DAPI, 5 mg ml−1).
Nu-clei were analysed on a PARTEC CA II flow cytometer (Chemunex, Maisons-Alfort, France) equipped with a 100-W mercury lamp (type HBO). Fluorescence at 455 nm was recorded as a func-tion of the DNA content. The DNA distribufunc-tion was analysed by using DPAC software on his-tograms generated from at least 104 nuclei. The dihaploid parental plants were used as external references to calibrate fluorescence scale.
Pollen viability was evaluated by staining pollen with fluorescein diacetate (FDA, 5 mg ml−1).
2.5. Isoenzyme analysis
Crude extracts were prepared using leaves from in vitro-grown plants [23]. A total of 20ml of each
sample was loaded on polyacrylamide gels (run-ning gel: 7.5% acrylamide+0.2% bisacrylamide; stacking gel: 4.5% acrylamide+0.12% bisacry-lamide), and electrophoresis was performed for 3 h at 4°C and 20 mA. Staining for esterases (E.C. 3.1.1.2) and peroxidases (E.C. 1.11.1.7) was done as described previously [28].
2.6. DNA analysis
Total DNA was obtained from frozen leaf tissue extracted with the DNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. PCR reactions leading to RAPD markers contained 30 ng DNA (0.5 ml of DNA sample), 10×Taq
1 U of Taq polymerase (Appligene) in a total volume of 25 ml. A total of 20 decamer
oligonu-cleotide primers from the kit AB-0320-1 (Fisher) and 14 primers previously described for potato by Xu et al. [29] (A02, A05, A10, A12, A16) and Baird et al. [30] (SC10-01, SC10-02, SC10-03, SC10-04, SC10-12, SC10-20, PBI-R005, ST1, ST2) were used. The thermal cycling profile, derived from Baird et al. [30], included 3-min denaturing at 92°C followed by 45 cycles of 92°C for 1 min, 37°C annealing for 1 min, and 72°C extension for 1 min. A final extension for 8 min at 72°C was the last step of the amplification program which was performed with a Touch Gene thermal cycler. Amplification products were electrophoresed onto 1.4% agarose gels. After staining with ethidium bromide, gels were pho-tographed on an UV box with Polaroid 665 films.
Polymorphic simple sequence repeat (SSR) markers in chloroplast genomes of Solanum
plants were recently described [31]. Several pairs of primers given by these authors (NTCP 6-1 and – 1 bis; NTCP 9-1 and – 1 bis; NTCP 12-1 and – 1 bis; Genaxis) have been used to distin-guish the chloroplast genomes of the parents and to characterise the ct genome type of the corre-sponding hybrids.
PCR amplification of chloroplast microsatel-lites was performed with the reaction as de-scribed above, primers excepted. The thermal cycling profile was that of Bryan et al. [31], in-cluding: 4 min denaturing at 94°C, followed by 30 cycles of 94°C for 1 min, annealing tempera-ture for 1 min, 72°C extension for 1 min and a final extension for 5 min at 72°C. Amplification products were electrophoresed onto 1.8% agarose gels which were stained and photographed as de-scribed above.
2.7. Determination of in 6itro resistance to
bacterial wilt
Two strains of R. solanacearum, G14* (race 1, biovar 3; isolated from Pelargonium asperum in Reunion) and PDT-5* (race 3, biovar 2; isolated from potato in Reunion), provided by CIRAD (Centre de Coope´ration Internationale en Recherche Agronomique pour le De´veloppement, Saint-Pierre, Reunion Island) were used to
inoc-ulate both parental lines and the somatic hy-brids. Cultures of R. solanacearum were routinely grown (24 h, 28°C) on basal medium, i.e. YPGA medium (yeast extract, 7 g l−1; peptone, 7 g l−1; glucose, 7 g l−1; agar, 15 g l−1; pH 7.2). Tests to evaluate sensitivity to bacterial wilt were per-formed on in vitro plants, using roots inoculated with suspension containing 107 colony forming units (cfu) per ml.
After cutting a part of the roots, the parental lines and somatic hybrids were soaked in the bacterial suspension for 30 min. Control plants were inoculated using sterile water. After inocu-lation, plants were placed in MS medium liquid, and exposed to 14 h day−1 illumination at 55 mmol m−2 s−1, 20°C and 60% relative humidity
in a controlled environment chamber. The tests for bacterial resistance were done by using 36 plants per clone, distributed over three replicates. Each replicate of 12 plants was distributed at random in the chamber. Plants were observed weekly and symptoms recorded using a disease index ranging from 0 to 4: 0=no wilted leaves, 1=up to 25% wilted, 2=up to 50% wilted, 3=
up to 75% wilted and 4=plants entirely wilted. Disease indices were calculated, according to Winstead and Kelman [32], as the ratio between the sum of the products of each disease number, divided by 4, and the corresponding number of plants and the total number of inoculated plants. Disease incidence was evaluated at d15 and d30 (15 days and 30 days after inoculation, re-spectively) by estimating the percentage of wilted plants that is with a disease index=4. Values of disease indices and disease incidence recorded at d15 and d30 were compared using a GStat test [33]. At d15, three inoculated but healthy looking plants belonging to the parental lines BF15 and
3. Results
3.1. Protoplast culture and plant regeneration
At 3 weeks after dilution with fresh VKM medium, dividing cells gave rise to hundreds of microcolonies, which developed rapidly into calli after the cultures were transferred onto the soli-dified growth medium. Early selection of the puta-tive somatic hybrids was based on the apparent difference in the cultural behavior of the parental and hybrid calli, particularly the ability of the latter to grow faster and regenerate early [34]. Therefore, 2 weeks later, only calli 2 – 3 mm in size were selected to be transferred onto the regenera-tion medium. Their growth was at least twice as rapid as that of the parental lines in the control cultures. After 5 weeks on the regeneration medium, some selected calli produced shoots with a frequency of one to five shoots per callus. Only one shoot was excised from the regenerating callus and multiplied by subculture on hormone-free MS medium. Finally, 55 of 322 selected calli produced shoots, representing a percentage of 17.0%.
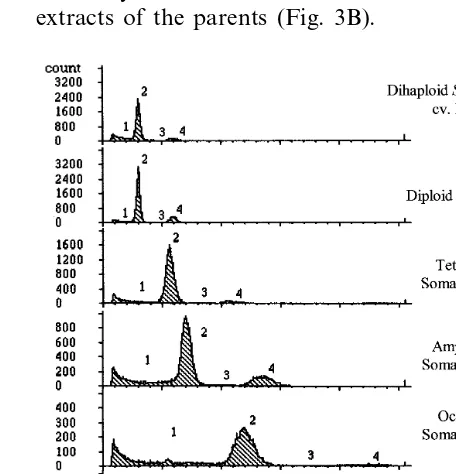
3.2. Ploidy le6el of the selected putati6e hybrids
The ploidy level was determined by comparing the position of dominant peaks corresponding to nuclei at G0-G1 phase of the cell cycle, between putative hybrid and parental plants (Fig. 1). Clear differences in peak position are shown in Fig. 1 between diploid parents and their somatic hybrids. The analysis of plants recovered from the fusion experiments showed that more than 80% were diploids. In total, ten plants were found to have a higher ploidy level. These plants were retained, as they were putative somatic hybrids. Among the selected plants, six (BP3, BP4, BP6, BP14, BP15, BP16) were tetraploids, two (BP1, BP8) mixo-ploids, one (BP9) amphiploid (\4×; 48) and one (BP13) octoploid (8×; 96). Chromosome counts made on root tips of a sample of the selected plants confirmed the ploidy level determined by flow cytometry.
3.3. Morphological analysis
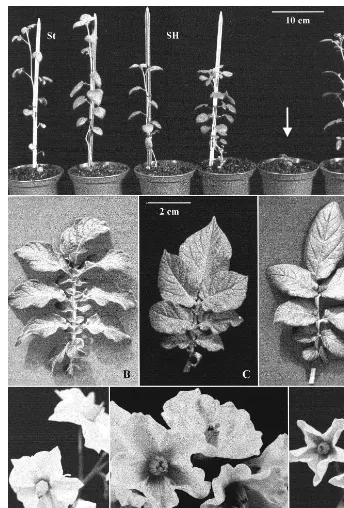
Both parental and putative somatic hybrid plants were grown up to maturity in the green-house. Most putative hybrid plants grew
vigor-ously and were larger than the parental plants. Their morphology was relatively homogeneous and intermediate between the parental lines, in-cluding leaves, flowers and tubers, all of which were larger than those of the parents (Fig. 2), except for the mixoploid clones having small leaves with irregular forms.
Pollen viability of putative hybrid flowers varied widely, ranging from 20 to 60% compared to 50 – 70% viable pollen in the parental plants.
3.4. Isoenzyme analysis
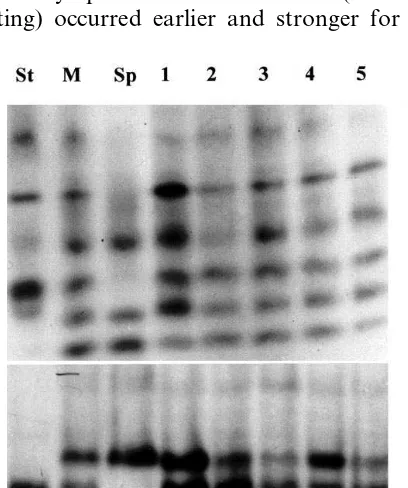
The hybrid nature of the selected putative hy-brids was confirmed by examining the elec-trophoretic patterns for esterases and peroxidases (Fig. 3). Each isoenzyme system revealed differ-ences between potato BF15 and S. phureja. They also distinguished somatic hybrids from the par-ents. The somatic hybrid pattern for esterases contained bands which were identical to those found in the mixed extracts of the parents (Fig. 3A). For peroxidase system in addition to the sum of the parental bands, the hybrid pattern showed additional bands, which were specifically relevant to the hybrid nature and not found in the mixed extracts of the parents (Fig. 3B).
Fig. 2. Plants:S.tuberosum(St),S.phureja(Sp) and four somatic hybrids (SH); the mixoploid is indicated by the arrow (A); leaves and flowers:S.tuberosum cv. BF15 (B, E),S.phureja(D, G) and their somatic hybrids (C, F).
3.5. DNA analysis
Out of 34 ten-mer primers tested, eight gave reproducible RAPD patterns showing polymor-phism between S. tuberosum cv. BF15 and S.
phureja. Three of them led to clear identification of the somatic hybrids. RAPD patterns obtained with the primer AB1-12 (5%-CCTTGACGCA-3%)
are given in Fig. 4A. Parental patterns can be
distinguished by DNA bands 500 – 1000 bp in size. The ten putative somatic hybrids showed similar patterns with specific parental bands and with an additional band (1.5 kb in size) which was absent from the parents.
The NTCP6 primers (5% -GGTTCGAATC-CTTCCGTC-3%and 5%
-GATTCTTTCGCATCTC-GATTC-3%) led to distinction between the two
specific ctDNA bands of 180 and 130 bp were amplified with the cultivated (as expected from Ref. [31]) and the wild species, respectively. All the somatic hybrids examined showed the ctDNA pattern of either one or the other parent. The ctDNA of potato was found in two somatic hybrids and the remaining eight hybrids possessed the S. phureja ct type (Fig. 4B).
Taking into account the intermediate morphol-ogy, the ploidy level and the analysis of nuclear and chloroplast genomes of the selected plants by examining the isoenzymes and DNA markers, it was concluded that the ten selected plants were somatic hybrids between S. tuberosum and S.
phureja.
3.6. E6aluation of resistance to bacterial wilt
S. tuberosum cv. BF15 was susceptible to both races, as 71.8 and 100% of plants were wilted 30 days (d30) after inoculation with races 1 and 3 strains, respectively, and disease indices reached 0.87 and 1.00 (Tables 1 and 2). Moreover, charac-teristic symptoms of bacterial wilt (necrosis and wilting) occurred earlier and stronger for the
di-Fig. 4. Electrophoresis profiles of PCR amplification prod-ucts. (A) RAPD patterns. DNA from S. tuberosum (St), S.
phureja(Sp) and their somatic hybrids BP3, BP4, BP6, BP15 and BP9 (lanes 1 – 5, respectively) was amplified using the primer AB1-12. Fractionation was on 1.4% agarose gels. The other five hybrids (not shown) led to patterns identical to the above hybrids. (B) Chloroplast microsatellite patterns. DNA from the same seven genotypes as above was amplified using NTCP6 primers. Fractionation was on 1.8% agarose gels. The other five hybrids (not shown) led to patterns identical toS.
phurejaL.: 100-bp DNA ladder (Biolabs).
haploid potato line than for the S. phurejaline.S.
phureja appeared to be tolerant to race 1 strain, showing a low disease indices (0.36) and no wilted plants at d30. S. phurejawas moderately suscepti-ble to race 3 strain with 50% of wilted plants at d30 but with a rather high disease indices (0.85) (Tables 1 and 2).
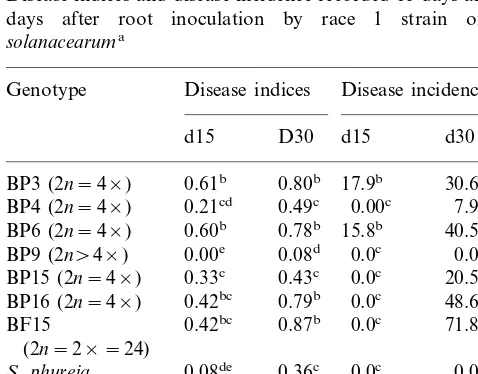
A total of six somatic hybrid plants, including five tetraploids and one amphiploid, were checked for resistance to bacterial wilt using race 1 and race 3 strains. Race 3 strain induced plant wilting earlier and with a higher frequency than did race 1 strain (Tables 1 and 2). Three tetraploid hybrids (BP3, BP6 and BP16) appeared moderately sus-ceptible to race 1 strain with 30.6 – 48.6% of wilted plants but their disease indices were not signifi-cantly different from that of the susceptible parent (0.78 – 0.80 versus 0.87). Two hybrids (BP4 and BP15) were less susceptible with 7.9 and 20.5% of wilted plants at d30, respectively, and their disease indices were not significantly different from that of the wild parent (Table 1). Only the amphiploid hybrid (BP9) was found at least as tolerant to race 1 strain as S. phureja since no wilted plants were recorded for both. However, its disease indices (0.08) was significantly lower than that of S.
phureja (0.36). All tetraploid hybrids were either more or at least similarly susceptible to race 3 strain compared to BF15 (Table 2). They were more susceptible than S. phureja, except BP4 which showed a similar wilting rate but a higher disease indices than the wild parent. Interestingly, the wilting rate of the amphiploid hybrid was low (2.8%) and not significantly different from zero. However, the disease indices (0.52) though signifi-cantly lower from that of S. phureja (0.85), would indicate that the hybrid was tolerant rather than resistant (Table 2).
Within roots of apparently healthy looking plants belonging either to both parental lines or to the amphiploid hybrid high populations (expressed as logarithms of cfu g−1fresh weight) were recov-ered, ranging from 7.27 to 7.93 whatever the clone or the strain. However, significant differences were recorded within stems of plants inoculated with race 1 strain between BF15 on the one hand (7.13) and S. phureja (3.74) and BP9 (3.20) on the other hand. For plants inoculated with race 3 strain, the bacterial populations within stems of both parental lines were significantly different although the difference was lower than one logarithmic unit. In contrast, the population within stem of the
Table 2
Disease indices and disease incidence recorded 15 days and 30 days after root inoculation by race 3 strain of R.
solanacearuma
aDisease indices is the weighted average of the disease
index. Disease index ranges from 0 to 4: 0=no wilted leaves, 1=up to 25% wilted, 2=up to 50% wilted, 3=up to 75% wilted and 4=plants entirely wilted. Disease incidence is the percentage of inoculated plants displaying a disease index of 4. Values followed by the same letter are not significantly different at P=0.05.
amphiploid hybrid was significantly reduced com-pared to both parents (Table 3).
4. Discussion
In this study, somatic hybrids between S.
tuberosum and S. phureja have successfully been produced by using electrofusion of protoplasts. The preliminary selection of putative hybrids was
Table 1
Disease indices and disease incidence recorded 15 days and 30 days after root inoculation by race 1 strain of R.
solanacearuma
Genotype Disease indices Disease incidence
d15 D30 d15 d30
aDisease indices is the weighted average of the disease
index. Disease index ranges from 0 to 4: 0=no wilted leaves, 1=up to 25% wilted, 2=up to 50% wilted, 3=up to 75% wilted and 4=plants entirely wilted. Disease incidence is the percentage of inoculated plants displaying a disease index of 4. Values followed by the same letter are not significantly different atP=0.05.
Table 3
Estimation of populations of R. solanacearum within roots and stems of apparently healthy looking plants, 15 days after root inoculation by race 1 or race 3 straina
Race 1 strain Race 3 strain
Root Stem Root Stem
aPopulations were expressed as the logarithm of cfu g−1
based on the difference in cultural behavior of calli, particularly callus growth and ability to re-generate shoots earlier. This selection method was effective, since 17% overall of regenerated plants were confirmed to be somatic hybrids. Obtaining a high number of somatic hybrids without using any selection system may be due to ploidy level and possibly to hybrid vigor of selected calli, which were able to regenerate early. Similar methods were successfully used to recover somatic hybrids of eggplant [34] and potato in particular [16,18,35].
The suitable characterisation of somatic hybrids is a prerequisite for the exploitation of protoplast fusion in crop improvement. Although numerous morphological markers are available to select and identify the hybrid plants, some of them are only expressed when the plants are grown to maturity. Therefore, neutral markers are needed to charac-terise putative hybrids in an early stage. In this study, both isozymes and RAPD markers were successfully used to identify and confirm the se-lected plants grown in vitro. RAPD markers were powerful for characterisation of nuclear genomes, and required only a very small quantity of plant material (generally 100 mg) for analysis [14]. Since they generally revealed dominant/null alleles [36], the presence of a polymorphic amplification product from each donor genotype is needed to confirm the presence of both genomes in putative hybrids. In this study, eight out of 34 primers used were necessary to generate specific patterns to identify somatic hybrids. In fact, since RAPD markers were expressed as dominant, it was re-quired for a single primer to generate at least one polymorphic amplification product from each par-ent to idpar-entify heterokaryons. In some cases, am-plification products present in the parents were not found in hybrids (data not shown). Conversely, in this study, primer AB1-12 amplified a novel band, 1.5 kb in size, which was observed in all the somatic hybrids analysed. Novel bands, not am-plified from any parent, have been reported in offspring of fungi, plants, insects and primates (reviewed in Ref. [37]). Non-parental bands might result from heteroduplex molecules formed be-tween allelic RAPD products, competition effects on primer sites in the genome [38] or/and so-maclonal variation within the somatic hybrids [30]. Although RAPD-based polymorphism assay can be an important tool for early identification of
potato interspecific fusion products, other marker assays which are not as sensitive to the above mentioned problems are currently available and may allow better characterisation of somatic hy-brids. Microsatellites, which are locus-specific and codominant markers, may be more effective for identification of nuclear polymorphism in plant genomes [39 – 41].
In this study, analysis of ctDNA indicated that among ten somatic hybrids obtained through elec-trofusion, eight possessed S. phureja ct type, and two potato ct type. Such apparently biased plastid transmission was described in somatic hybrids be-tween tomato and S. lycopersicoides (partially in-compatible with potato) [41] and also in somatic hybrids of eggplant with its wild relatives [34,42]. In our experiments, the small number of the so-matic hybrids recovered did not allow us to deter-mine whether chloroplast transmission was significantly biased or not.
The response of the parental lines to inoculation with R. solanacearum varied with the strain that was used. S. tuberosum cv. BF15 was found more susceptible to race 3 than race 1 strains. The wild species, S. phureja, used as fusion partner in this study, appeared tolerant to race 1 strain and mod-erately susceptible to race 3 strain under our ex-perimental conditions. This apparent difference in susceptibility to bacterial wilt may be due to dif-ferences in bacterial colonisation of stems: popula-tions of race 1 strain in BF15 stem were more than 107 cfu mg−1 FW whereas there were only a few bacteria inS. phurejastem. In contrast, more than 106cfu mg−1FW were recovered from S. phureja stems inoculated with race 3 strain. These results were in agreement with those for tomato regarding the relationship between cultivar resistance or tol-erance to bacterial wilt and density of bacterial population inside the stems [2,43]. Restricted colonisation of resistant plants byR.solanacearum
was recently reported to be related to the produc-tion of tyloses and other non-specific physical barriers [44]. Moreover, it was shown that potato resistance against bacterial wilt was highly en-hanced when genes encoding for both biotic and abiotic resistance were combined together [45]. Resistance to race 1 strain was reported to be more stable than resistance to race 3 strain [46], but was overcome by high temperature [7,12].
(BP9) (\48 chromosomes) was found more tol-erant to race 1 and race 3 strains than the wild parent, S. phureja. This novel gene combination is probably due to the association of one copy of dihaploid potato (2×) with one copy plus a few chromosomes of S. phureja genome (\2×), probably harboring the resistance genes, and giv-ing a putative gene dosage with a higher expres-sion of tolerance in the amphiploid hybrid. This observation agrees with the fact that the level of bacterial colonisation of the amphiploid hybrid stems was found either similar to (race 1) or lower than (race 3) that of the wild species, S.
phureja.
In view of results obtained in this study, to-gether with those from others for introgression of bacterial resistance in particular [19,47], so-matic fusion is a valid means of introducing traits of resistance from wild into cultivated potatoes, and to complement and supplement conventional breeding methods. Moreover, so-matic fusion provided a substantial time saving over conventional sexual hybridisation [48]. As described for viruses [14,15] or for fungi [17], traits of resistance are expressed in somatic hy-brids with some variation in expression that could be due to the complex polygenic inheri-tance of resisinheri-tance [14]. It has also been reported regarding late blight of potatoes (P. infestans) that somatic hybrids are less resistant than the wild parental species of the fusion whereas their F1 progeny could express the full resistance of the resistant parent [47]. Two somatic hybrid clones of potato with S. phureja obtained in this study had the level of tolerance to race 1 and race 3 strains of R. solanacearum, which was as high as that of the wild species. Interestingly, the highest level of tolerance was found in an am-phiploid hybrid clone. Further detailed evalua-tion of the somatic hybrids in field condievalua-tions is needed to confirm the results of in vitro tests for bacterial tolerance, and also to validate their ex-ploitation in potato breeding programs.
Acknowledgements
This study was conducted as part of the doc-toral research by the first author who would like to thank the Conseil Ge´ne´ral de la Re´union for awarding her a fellowship.
References
[1] A.C. Hayward, J.G. Elphinstone, D. Caffier, J. Janse, E. Stefani, E.R. French, A.J. Wright, Round table on bacterial wilt (brown rot) of potato, in: P. Prior, C. Allen, J. Elphinstone (Eds.), Bacterial Wilt Disease, Molecular and Ecological Aspects, Springer, Berlin, 1998, pp. 420 – 430.
[2] A.C. Hayward, The hosts ofPseudomonas solanacearum, in: A.C. Hayward, G.L. Hartman (Eds.), Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas solanacearum, CAB International, Wallingford, UK, 1994, pp. 9 – 23.
[3] V. Grimault, P. Prior, Bacterial wilt resistance in tomato associated with tolerance of vascular tissues to Pseu
-domonas solanacearum, Plant Pathol. 42 (1993) 589 – 594. [4] E.R. French, R. Anguiz, F.P. Aley, The usefulness of potato resistance toRalstonia solanacearum, for the inte-grated control of bacterial wilt, in: P. Prior, C. Allen, J. Elphinstone (Eds.), Bacterial Wilt Disease, Molecular and Ecological Aspects, Springer, Berlin, 1998, pp. 381 – 385.
[5] E.R. French, Strategies for integrated control of bacte-rial wilt of potatoes, in: A.C. Hayward, G.L. Hartman (Eds.), Bacterial Wilt: The Disease and Its Causative Agent, Pseudomonas solanacearum, CAB International, Wallingford, UK, 1994, pp. 199 – 207.
[6] I.W. Buddenhagen, Bacterial wilt revisited, in: G.J. Pers-ley (Ed.), Bacterial Wilt Disease in Asia and the South Pacific. In: ACIAR Proceedings, vol. 13, 1986, pp. 126 – 143.
[7] A.C. Hayward, Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum, Annu. Rev. Phytopathol. 29 (1991) 65 – 87.
[8] C.A. Lopes, A.M. Quezado-Soares, J.A. Buso, P.E. Melo, Breeding for resistance to bacterial wilt of pota-toes in Brazil, in: P. Prior, C. Allen, J. Elphinstone (Eds.), Bacterial Wilt Disease, Molecular and Ecological Aspects, Springer, Berlin, 1998, pp. 290 – 293.
[9] J.G. Hawkes, The Potato. Evolution, Biodiversity and Genetic Resource, Belhaven Press, London, 1990, 259 pp.
[10] L. Sequeira, P.R. Rowe, Selection and utilisation of
Solanum phurejaclones with high resistance to different strains ofPseudomonas solanacearum, Am. Potato J. 46 (1969) 451 – 462.
[11] P.R. Rowe, L. Sequeira, Inheritance of resistance to
Pseudomonas solanacearum in Solanum phureja, Phyto-pathology 60 (1970) 1499 – 1501.
[12] E.R. French, L. De Lindo, Resistance to Pseudomonas solanacearum in potato: specificity and temperature sen-sitivity, Phytopathology 72 (1982) 1408 – 1412.
[13] N. Grimley, P. Hanson, Genetics of plant resistance to bacterial wilt: round table report, in: P. Prior, C. Allen, J. Elphinstone (Eds.), Bacterial Wilt Disease, Molecular and Ecological Aspects, Springer, Berlin, 1998, pp. 263 – 266.
[14] V.M. Rokka, Y.S. Xu, J. Kankila, A. Kuusela, S. Pulli, E. Pehu, Identification of somatic hybrids of dihaploid
RAPD patterns and assessment of disease resistance of the hybrids, Euphytica 80 (1994) 207 – 217.
[15] J.P.T. Valkonen, V.M. Rokka, Combination and expres-sion of two virus resistance mechanisms in interspecific somatic hybrids of potato, Plant Sci. 131 (1998) 85 – 94. [16] J. Preiszner, A. Feher, O. Veisz, J. Sutka, D. Dudits, Characterization of morphological variation and cold resistance in interspecific somatic hybrids between potato (Solanum tuberosum L.) and S. bre6idens Phil.,
Euphytica 57 (1991) 37 – 49.
[17] W.M. Mattheij, K. Eijlander, J.R.A. de Koning, K.M. Louwes, Interspecific hybridization between the culti-vated potato (Solanum tuberosum subspecies tuberosum
L. and the wild species S. circaeifolium subsp. circaei
-foliumBitter exhibiting resistance to Phytophtora infes
-tans (Mont.) de Bary and Globodera pallida (Stone) Behrens, Theor. Appl. Genet. 83 (1992) 459 – 466. [18] I. Serraf, D. Sihachakr, G. Ducreux, S.C. Brown, M.
Allot, N. Barghi, L. Rossignol, Interspecific somatic hybridization in potato by protoplast electrofusion, Plant Sci. 76 (1991) 115 – 126.
[19] L.T. Laferriere, J.P. Helgeson, C. Callen, Fertile
Solanum tuberosum+S.commersonii somatic hybrids as sources of resistance to bacterial wilt caused byRalsto
-nia solanacearum, Theor. Appl. Genet. 98 (1999) 1272 – 1278.
[20] L.P. Pijnacker, M.A. Ferwerda, K.J. Pulter, S. Roest, Elimination ofSolanum phurejanucleolar chromosomes in S. tuberosum+S. phureja somatic hybrids, Theor. Appl. Genet. 73 (1987) 878 – 882.
[21] L.P. Pijnacker, M.A. Ferwerda, K.J. Pulte, J.G. Schaart, Chromosome elimination and mutation in tetraploid somatic hybrids of Solanum tuberosum and S. phureja, Plant Cell Rep. 8 (1989) 82 – 85.
[22] T. Murashige, F. Skoog, A revised medium for rapid growth and bioassays with tobacco tissue cultures, Phys-iol. Plant. 15 (1962) 473 – 597.
[23] M.H. Chaput, D. Sihachakr, G. Ducreux, D. Marie, N. Barghi, Somatic hybrid plants produced by electrofusion between dihaploid potatoes: BF15 (H1), Aminca (H6) and Cardinal (H3), Plant Cell Rep. 9 (1990) 411 – 414. [24] E.M. Frearson, J.B. Power, E.C. Cocking, The isolation,
culture and regeneration of Petunia leaf protoplasts, Dev. Biol. 33 (1973) 130 – 137.
[25] D. Sihachakr, R. Haicour, I. Serraf, E. Barrientos, C. Herbreteau, G. Ducreux, L. Rossignol, V. Souvan-navong, Electrofusion for the production of somatic hybrid plants of Solanum melongena L. and Solanum khasianumcv. Clark, Plant Sci. 57 (1988) 215 – 223. [26] H. Binding, R. Nehls, O. Schieder, S.K. Sopory, G.
Wenzel, Regeneration of mesophyll protoplasts isolated from dihaploid clones of Solanum tuberosum, Physiol. Plant. 43 (1978) 52 – 54.
[27] G. Morel, R.-H. Wetmore, Fern callus tissue culture, Am. J. Bot. 38 (1951) 141 – 143.
[28] C.R. Shields, T.P. Orion, C.W. Stuber, An outline of general resource needs and procedures for the elec-trophoretic separation of active enzymes from plant tissue, in: S.D. Tanskley, T.P. Orion (Eds.), Isoenzymes in Plant Genetics and Breeding, Part A, Elsevier, Am-sterdam, 1983, pp. 443 – 516.
[29] Y.S. Xu, M.S. Clark, E. Pehu, Use of RAPD markers to screen somatic hybrids betweenSolanum tuberosumand
S.bre6idens, Plant Cell Rep. 12 (1993) 107 – 109.
[30] E. Baird, S. Cooper-Bland, R. Waugh, M. DeMamie, W. Powell, Molecular characterisation of inter- and intra-specific somatic hybrids of potato using randomly amplified polymorphic DNA (RAPD) markers, Mol. Gen. Genet. 233 (1992) 469 – 475.
[31] G.J. Bryan, J.M. McNicoll, G. Ramsay, R.C. Meyer, W.S. De Jong, Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants, Theor. Appl. Genet. 99 (1999) 859 – 867.
[32] N.N. Winstead, A. Kelman, Inoculation techniques for evaluating resistance toPseudomonas solanacearum, Phy-topathology 42 (1952) 628 – 634.
[33] R.R. Sokal, F.J. Rolf, The principles and practice of statistics in biological research, in: Biometry, Freeman, San Francisco, 1969, 776 pp.
[34] D. Sihachakr, M.C. Daunay, I. Serraf, M.H. Chaput, I. Mussio, R. Haicour, L. Rossignol, G. Ducreux, Somatic hybridisation of eggplant (Solanum melongena L.) with its close and wild relatives, in: Y.P.S. Bajaj (Ed.), So-matic Hybridisation in Crop Improvement I. In: Bio-technology in Agriculture and Forestry, vol. 27, Springer, Berlin, 1994, pp. 255 – 278.
[35] I. Serraf, S. Tizroutine, M.H. Chaput, M. Allot, I. Mussio, D. Sihachakr, L. Rossignol, G. Ducreux, Pro-duction and characterisation of intergeneric somatic hy-brids through protoplast electrofusion between potato (Solanum tuberosum) and Lycopersison pennellii, Plant Cell. Tissue Org. Cult. 37 (1994) 137 – 144.
[36] J.G.K. Williams, A.R. Kubelik, K.J. Livak, J.A. Rafal-ski, S.V. Tingey, DNA polymorphisms amplified by arbitrary primers are useful as genetic markers, Nucleic Acids Res. 18 (1990) 6531 – 6535.
[37] A. Reineke, P. Karlovsky, C.P.W. Zebitz, Suppression of randomly primed polymerase chain reaction products in heterozygous diploids, Mol. Ecol. 8 (1999) 1449 – 1455.
[38] T. Richardson, S. Cato, J. Ramser, G. Kahl, K. Weis-ing, Hybridization of microsatellites to RAPD: a new source of polymorphic markers, Nucleic Acids Res. 23 (1995) 3798 – 3799.
[39] J. Provan, W. Powell, R. Waugh, Microsatellite analysis of relationships within cultivated potato (Solanum tuberosum), Theor. Appl. Genet. 92 (1996) 1078 – 1084. [40] D. Milbourne, R. Meyer, J.E. Bradshaw, E. Baird, N.
Bonar, J. Provan, W. Powell, R. Waugh, Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato, Mol. Breed. 3 (1997) 127 – 136.
[41] A. Levi, B.L. Ridley, K.C. Sink, Biased organelle trans-mission in somatic hybrids of Lycopersicon esculentum
and Solanum lycopersicoides, Curr. Genet. 14 (1988) 177 – 182.
[42] M.C. Daunay, M.H. Chaput, D. Sihachakr, M. Allot, F. Vedel, G. Ducreux, Production and characterisation of fertile somatic hybrids of eggplant (Solanum melongena
[43] V. Grimault, B. Ge´lie, M. Lemattre, P. Prior, J. Schmit, Comparative histology of resistant and susceptible tomato cultivars infected byPseudomonas solanacearum, Physiol. Mol. Plant Pathol. 44 (1994) 105 – 123. [44] P. Prior, S. Bart, S. Leclercq, A. Darrasse, G. Anais,
Resistance to bacterial wilt in tomato as discerned by spread of Pseudomonas (Burkolderia) solanacearum in the stem tissues, Plant Pathol. 45 (1996) 720 – 726. [45] J.G. Elphinstone, Inheritance of resistance to bacterial
diseases, in: J.E. Bradshaw, G.R. Mackay (Eds.), Potato Genetics, CAB International, Wallingford, UK, 1994, pp. 429 – 446.
[46] G.L. Hartman, J.G. Elphinstone, Advances in the con-trol ofPseudomonas solanacearumrace 1 in major food
crops, in: A.C. Hayward, G.L. Hartman (Eds.), Bacte-rial Wilt: The Disease and Its Causative Agent, Pseu
-domonas solanacearum, CAB International, Wallingford, UK, 1994, pp. 157 – 177.
[47] L.T. Laferriere, J.P. Helgeson, C. Allen, Solanum tuberosum-S. commersonii somatic hybrids are resistant to brown rot caused by Ralstonia solanacearum, in: P. Prior, C. Allen, J. Elphinstone (Eds.), Bacterial Wilt Disease, Molecular and Ecological Aspects, Springer, Berlin, 1998, pp. 316 – 320.
[48] S. Millam, L.A. Payne, G.R. Mackay, The integration of protoplast fusion-derived material into a potato breed-ing programme — a review of progress and problems, Euphytica 85 (1995) 451 – 455.