J. Bacteriol. Research Article
D,L-endopeptidase activity at lateral cell wall is essential for Bacillus subtilis cell proliferation.
I Putu Suparthana2, Masayuki Hashimoto1, Matsushima Hiroaki2, and
Junichi Sekiguchi2,*.
1
Department of Biological Sciences, Graduate Schools of Science and Engineering, Tokyo
Metropolitan University, Tokyo, Japan.2 Department of Applied Biology, Faculty of Textile Science and Technology, Shinshu University, Ueda, Japan.
Running title: DLEPase is essential for B. subtilis proliferation Section: Genetics and Molecular Biology
Abstract
Most bacterial cells possess peptidoglycan layer(s), which is an exoskeleton of bacterial cells. To proliferate the bacterial cells, the peptidoglycan must change the shape, and have a dynamic structure. Thus, not only synthesis but also disassemble of peptidoglycan are important for the cell proliferation. In Bacillus subtilis, a double mutant of lytE and cwlO,both encoding D,L-endopeptidase known asautolysin, showed
synthetic lethality and defected in cell elongation. Furthermore, the synthetic lethality is caused by lack of D,L-endopeptidase activity at the lateral cell wall. In this study, to
investigate the function of LytE and CwlO, we constructed some chimeric enzymes, which locate at cellular sidewall and exhibit various autolysin activities except
D,L-endopeptidase, and demonstrated whether the chimeric enzymes can suppress the
synthetic lethality or not. As the results, all chimeric enzymes except with M23 domain of CwlP, which is a D,D-endopeptidase, did not suppress the lytE cwlO synthetic
lethality. In addition, the suppression by M23 domain of CwlP was caused by its intense activity. Therefore, we concluded that D,L-endopeptidase activity at lateral cell wall is
essential for Bacillus subtilis cell proliferation. The function of D,L-endopeptidase
activity at lateral cell wall was discussed.
(Introduction)
Bacterial cells are generally covered with peptidoglycan (PG), and the PG determines shape of the cell. The PG forms a mesh-like structure, which is comprised of polysaccharide chains and peptide bridges (Fig. 1). This mesh-like structure behaves as an exoskeleton of bacterial cells, and the cells can tolerate various environmental stresses with PG sacculus. Therefore, PG is essential for cell viability except under special conditions. During bacterial cell proliferation, the cells grow bigger and divide into two daughter cells. While cells growing, the PG sacculus covering the cell have to change the shape. Therefore, the PG sacculus must be a dynamic structure, and not only synthesis but also reassemble of the structure is important. PG synthesis by
penicillin-binding proteins has been well studied, but only few studies have described the PG disassemble by cell wall degrading enzymes for proliferation. It was reported recently that double disruption of lytE cwlO in Bacillus subtilis shows synthetic lethal, and it is only known that defect of cell wall degrading enzyme genes shows lethality to date (Bisicchia 2007).
LytE, LytF, CwlS and CwlO are belonging to D,L-endopeptidase (DLEPase), which
hydrolyze the linkage of D -γ-glutamyl-meso-diaminopimelic acid in PG (Ishikawa 1998,
Yamaguchi 2004). LytE, LytF and CwlS were detected at cellular poles and septa, and some mutants disrupted these enzymes showed chain-like cell shapes, and were defect in cell separation (Yamamoto 2003, Fukushima 2006). Besides, LytE and CwlO were detected at cellular sidewall and a mutant of these enzymes was lethal and defect in cell elongation (Carballido-Lopez 2006, Bisicchia 2007, Hashimoto 2012). These results led us propose that the cellular localization of these DLEPases determine their function for cell morphogenesis. In support of this proposition, the lytE cwlO synthetic lethality was suppressed by some chimeric enzymes of these DLEPases only which were active and localized at cellular sidewall (Hashimoto 2012). These results revealed that lytE cwlO synthetic lethal is caused by lack of DLEPase activity at lateral cell wall. Taken together, it was strongly suggested that the DLEPase activity at lateral cell wall involves in cell elongation (Bisicchia 2007, Hashimoto 2012).
B. subtilis has thick PG layers in the cell wall, and three distinct part in the
thick layers was observed by electron microscopic observation (Merad 1989, Graham 1994). In addition, pulse labeling study demonstrated that newly synthesized PG was released into medium with a delay of more than one generation (Pooley 1976). These results suggested that the inner zone of the thick PG layers is newly synthesized layers, and the outer zone consists old degraded layers. Furthermore, up to half of the
preexisting PG is released from B. subtilis cell in one generation (Reith 2011). Taken together of these results, an inside-to-outside growth model for PG layers formation is suggested. In this model, outer zone of the thick PG layers have to degrade to make a space for newly synthesized PG layers at inner zone. As described above, DLEPase activity at lateral cell wall is essential for cell growth, and lytE cwlO mutant defected in cell elongation. These imply that function of LytE and CwlO is degradation of outer old PG layer at cellular sidewall. In such a case, the DLEPase activity at lateral cell wall might be able to substitute by the other cell wall lytic activity. Then, to investigate the function of LytE and CwlO for PG dynamic structure, we examined whether the DLEPase activity is essential for cell proliferation or is able to substitute by the other activity.
MATERIALS AND METHODS
Materials and growth conditions. Strains and plasmids used in this study are listed in
Table 1, and primers are in Table S1. Escherichia coli and Bacillus subtilis were grown
in the following concentrations: ampicillin, 50 µg/ml; spectinomycin, 50 µg/ml; chloramphenicol, 5 µ g/ml; xylose, 1%.
Strain constructions. DNA manipulation and E. coli transformation were performed
using standard methods (Sambrook 1989). B. subtilis transformation was performed by conventional transformation procedures (Anagnostopoulos 1961). For each construction of strain expressing LytE chimeric enzyme, a plasmid was constructed by using
pCA6FL as the vector. A fragment encoding N-terminal domain of LytE and a fragment encoding each catalytic domain of various autolysins were amplified by PCR with B.
subtilis chromosome as a template. Except for construction of point mutated
LytE-CwlP(M23), pQEM23-H1580Q for pCA-NeCcwlP(M23)1580, pQEM23-D1584N for pCA-NeCcwlP(M23)1584 and pQEM23-H1662Q for pCA-NeCcwlP(M23)1620 were used as the templates. Sets of primers for each PCR amplification are below: LytE; LytE-19 and LytE-20, LytD3; LytD-3 and LytD-2, LytD4; LytD-4 and LytD-2, CwlQ; YjbJ-1 andYjbJ-2, CwlP(SLT); YomI-1 and YomI-2, LytC; LytC-1 and LytC-2, CwlK; CwlK-1 and CwlK-2, LytH; LytH-1 and LytH-2, CwlP(M23); YomI-3 and YomI-4. The amplified fragments encoding N-terminal domain of LytE and each catalytic domain were used as template for second PCR with proper primers for amplification of their connected fragments. The amplified fragments encoding the chimeric enzymes were treated with KpnI and BamHI and cloned into pCA6FL treated with the same restriction enzymes. For construction of pCA-NeClytD3 and pCA-NeClytD4, however, KpnI and
XbaI were used for the cloning. To construct strains expressing a chimeric enzyme, B.
subtilis PxOd harboring Pxyl-cwlO (Spc) was transformed with the obtained plasmid
(Table 1). The all constructed genetic structures were confirmed by PCR.
Western blotting and zymography. Western blotting and zymography were done as
described previously (Hashimoto et al 2012). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and zymography were performed with xxx% polyacrylamide gels.
Immunofluorescent microscopy (IFM). Samples for immunofluorescence microscopy
(IFM) were prepared as described previously (Hashimoto 2012). Fluorescence microscopy was performed with an AxioImager M1 microscope, a
Results
Strains expressing chimeric enzyme. To investigate the DLEPase activity at lateral
cell wall whether able to substitute by other cell wall lytic activity or not, we
constructed strains expressing a chimeric enzyme, which catalytic domain of LytE was substituted by that of the other autolysins. The chimeric enzyme is comprised of cell wall binding domain (N-terminal domain) of LytE at N-terminus, a catalytic domain of other autolysins, and 6xFLAG at C-terminus. Since the cell wall binding domain of LytE localizes at cellular poles, septa and lateral cell wall, the chimeric enzymes are recruited to there. In addition, 6xFLAG is fused at C-terminus to detect these chimeric enzymes with anti FLAG antibody on Western blotting and IFM. A promoter of these chimeric enzymes is intact lytE promoter. On the other hand, that of cwlO substituted by xylose inducible promoter (Pxyl). Therefore, it is able to observe by culturing the strains without xylose that whether the chimeric enzyme suppresses the lytE cwlO lethality or not. The catalytic domains comprised to chimeric enzymes are shown in Fig. 1. Catalytic domains of eight autolysins were used for construction of the chimeric enzymes. Consequently, all groups except for D,D-carboxypeptidase were covered by
some chimeric enzymes.
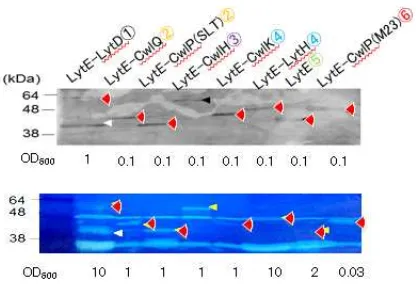
Expression and activity of chimeric enzymes. Fig. 2 shows results of Western blotting
and zymography of the strains expressing chimeric enzymes (Fig. 2). Samples applied to them were cultured with xylose to induce CwlO expression. As the result of Western blotting, signals for all chimeric enzymes were detected at proper positions to their predicted molecular sizes. For NLytECLytD3 and NLytECLytD4, however, signals for lower molecular size than predicted were also detected. A part of these chimeric enzymes were likely proteolyzed. On the zymography, clear zones corresponding to all signals on Western blotting were detected. Since clear zones for the lower size of NLytECLytD3 and NLytECLytD4 were also detected, these chimeric enzymes may be proteolyzed at
N-terminal. These results demonstrated that all chimeric enzymes were expressed as active form, although a part of them were proteolyzed.
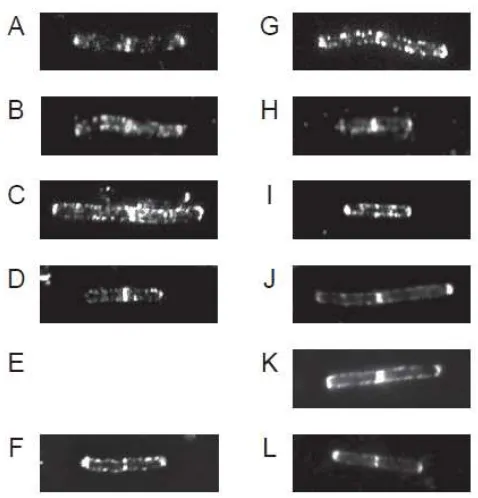
Cellular localization of chimeric enzymes. To confirm the function of cell wall bind
domain of LytE comprising the chimeric enzymes, cellular localization of these chimeric enzymes were observed by IFM (Fig. 3). As the result, all chimeric enzymes were detected at cellular sidewall, septa and poles similar with LytE-6xFLAG.
detected at lateral cell wall contain their catalytic domain, even though a part of them were proteolyzed.
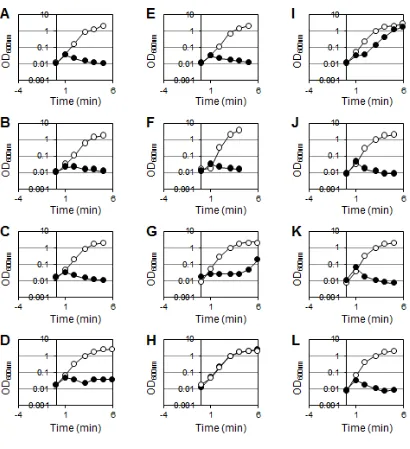
Suppression of lytE cwlO synthetic lethality by the chimeric enzymes. As described
above, since the promoter for cwlO in the strains expressing chimeric enzyme is substituted by Pxyl, functionality of the chimeric enzymes is able to examine by
depletion of CwlO. To investigate whether DLEPase activity at lateral cell wall is able to substitute by other cell wall lytic activity or not, the strains expressing the chimeric enzyme were cultured with or without 1% xylose (Fig. 4). In the presence of xylose, all strain showed similar growth with wild type. In the absence of xylose, on the other hand, growth of all strains except for NLytECCwlP(M23) were stopped, thus these chimeric
enzymes did not suppress the lytE cwlO synthetic lethality. However, NLytECCwlP(M23) cultured without xylose apparently grew, although the growth was slightly slower than wild type. Therefore, NLytECCwlP(M23), which contains a catalytic domain of
D,D-endopeptidase (DDEPase), is able to suppress the lytE cwlO synthetic lethality.
However, the activity of NLytECCwlP(M23) was quite higher than that of LytE on
zymography (Fig. 2). Furthermore, specific activities of purified CwlP(M23) catalytic domain was 28 times higher than that of LytE (Fig. S1). Hence, there is a possibility that NLytECCwlP(M23) suppressed the synthetic lethality as non-specific function by the higher activity. Then, to assess whether the suppression by xxx was due to the higher activity or not, point-mutated chimeric enzymes of CwlP (M23) were used for the suppression assay (Fig. 4). The activities of chimeric enzymes with point-mutated CwlP(M23) on zymography were comparable to LytE-6xFLAG (Fig. 2). However, the point-mutated chimeric enzymes did not suppress the lytE cwlO synthetic lethality. Thus, DDEPase activity comparable with LytE activity could not suppress the lytE
cwlO synthetic lethality. These results indicated that the suppression by NLytECCwlP(M23)
was non-specific suppression due to the intense activity. Taken together these results, we concluded that DLEPase activity at lateral cell wall is essential for cell proliferation.
Discussion
In this study, we demonstrated that essential DLEPase activity at lateral cell wall is not able to substitute by the other cell wall lytic activity, although NLytECCwlP(M23) non-specifically suppressed the lytE cwlO synthetic lethality due to the intense activity. Nevertheless, all constructed chimeric enzymes have cell wall lytic activity and
The synthetic lethality of lytE cwlO was suppressed by only NLytECCwlP(M23), which has DDEPase activity. As described above, however, this chimeric enzyme exhibited intense activity on zymography, and purified M23 domain of CwlP showed 28 times higher specific activity than that of LytE (Fig. 2, Fig. S1). Then, the following three possibilities were considered: 1) the essential DLEPase activity at lateral cell wall is able to replace with DDEPase activity, 2) CwlP(M23) has also DLEPase activity, and 3) NLytECCwlP(M23) can suppress by non-specifically due to the intense activity. Since CwlP(M23) is the only enzyme known as DDEPase in B. subtilis to date, we cannot demonstrate the suppressivity by using other DDEPase from B. subtilis. To examine it, we constructed a chimeric enzyme by using PBP8 from E. coli, which has DDEPase activity (Romeis 1994). However, the chimeric enzyme with PBP8 did not show any cell wall lytic activity on zymography (data not shown). The second possibility has been denied by Sudiarta and co-workers (Sudiarta 2010). They compared the activity of purified M23 domain of CwlP with that of LytF as a certain DLEPase, however, the M23 domain did not show any DLEPase activity. This conclusion is supported by the result, that the M23 domain did not degrade cell wall from Streptococcus aureus (Sudiarta 2010). The linkage of D -γ-glutamyl-meso-diaminopimelic acid, which is
cleaved by DLEPase, is common in both PG from B. subtilis and S. aureus, but the structures of peptide cross linkage, which is cleaved by DDEPase, are different in their PG. If the M23 domain had a DLEPase activity, the enzyme should degrade cell wall from S. aureus. To assess the last possibility, we constructed some point-mutated NLytECCwlP(M23). According to Sudiarta et al, point mutation at H1580Q, D1584N and H1662Q of the M23 domain of CwlP showed 6.1%, 5.6% and 8.9% activity to the intact enzyme on zymography, respectively (Sudiarta 2010). Activities of the point-mutated NLytECCwlP(M23) decreased similar with the previous report, and are comparable with LytE-6xFLAG (Fig. 2). However, these point mutated chimeric enzyme did not suppress the synthetic lethality, although intact NLytECCwlP(M23) did. Thus, it is strongly suggested that the suppression by NLytECCwlP(M23) was non-specific suppression by the intense cell wall degrading activity.
DLEPase activity played the role to degrade the outer zone of PG layers, the activity should be able to substitute by the other cell wall lytic activity. Thus, the results in this study imply that the essential function of DLEPase activity at cellular sidewall is not to degrade the outer zone of PG layers, and it may has a specific function for PG dynamics. Carballido-Lopez et al reported that LytE has some interaction to MreBH, an MreB homologue (Carballido-Lopez). MreBH and the other MreB homologues colocalize in helical manner at lateral sidewall as a scaffold of PG synthesis enzymes. Maybe the DLEPase activity directly involved in PG biosynthesis at inner zone of PG layers. Only DDEPase activity was substitutable to the DLEPase activity, although the intense DDEPase activity is necessary. The cleavage site of DDEPase is the linkage between
D-Ala and diaminopimelic acid, which comprises the cross-linked peptide side chains of
B. subtilis PG. DDEPase can potentially suppress the synthetic lethality, since the
position of site cleaved by DDEPase is more outer from polysaccharide chain than that of DLEPase. To understand the function of essential DLEPase activity at lateral cell wall, further analyses are necessary.
Acknowledgements
We would like to thank the members of our group, particularly Dr. Hiroki Yamamoto and Dr. Ryoichi Arai, for the helpful advice and discussion. This work was supported by Grants-in-Aid for Scientific Research (B) (19380047) and (A) (22248008) (JS), and the Program for Dissemination of Tenure-Track System funded by the
Ministry of Education and Science, Japan (MH).
References
Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in
Bacillus subtilis. J. Bacteriol. 81:741-746.
Bisicchia, P., D. Noone, E. Lioliou, A. Howell, S. Quigley, T. Jensen, H. Jarmer,
and K. M. Devine. 2007. The essential YycFG two-component system controls cell wall
metabolism in Bacillus subtilis. Mol. Microbiol. 65:180-200.
Carballido-Lopez, R., A. Formstone, Y. Li, S. D. Ehrlich, P. Noirot, and J.
Errington. 2006. Actin homolog MreBH governs cell morphogenesis by localization of
the cell wall hydrolase LytE. Dev. Cell. 11:399-409.
Fukushima, T., A. Afkham, S. Kurosawa, T. Tanabe, H. Yamamoto, and J.
Sekiguchi. 2006. A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis. J. Bacteriol.
Graham, L. L., and T. J. Beveridge. 1994. Structural differentiation of the Bacillus
subtilis 168 cell wall. J. Bacteriol. 176:1413-1421.
Hashimoto M, Ooiwa S, and Sekiguchi J. 2012. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral
cell wall. J. Bacteriol. 194:796-803.
Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol.
180:2549-2555.
Merad, T., A. R. Archibald, I. C. Hancock, C. R. Harwood, and J. A. Hobot. 1989. Cell wall assembly in Bacillus subtilis visualization of old and new wall material by electron microscopic examination of samples stained selectively for teichoic acid and
teichuronic acid. J. Gen. Microbiol. 135:645-655.
Pooley, H. M. 1976. Layered distribution, according to age, within cell wall of Bacillus
subtilis. J. Bacteriol. 125:1139-1147.
Reith J and Mayer C. 2011. Peptidoglycan turnover and recycling in Gram-positive
bacteria. Appl. Microbiol. Biotechnol. 92:1-11.
Romeis T, Höltje JV. 1994. Penicillin-binding protein 7/8 of Escherichia coli is a
DD-endopeptidase. Eur. J. Biochem. 224:597-604.
Sambrook, J., E. F. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 146:249-262.
Sudiarta IP, Fukushima T, and Sekiguchi J. 2010. Bacillus subtilis CwlP of the SP-β prophage has two novel peptidoglycan hydrolase domains, muramidase and
cross-linkage digesting DD-endopeptidase. J. Biol. Chem. 285:41232-43.
Yamaguchi, H., K. Furuhata, T. Fukushima, H. Yamamoto, and J. Sekiguchi. 2004.
Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named
cwlO), and the enzymatic properties of its encoded protein. J. Biosci. Bioeng.
98:174-181.
Yamamoto, H., S. Kurosawa, and J. Sekiguchi. 2003. Localization of the vegetative
cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall bound or extracellular proteases. J. Bacteriol.
185:6666-6677.
Yamamoto, H., M. Hashimoto, Y. Higashitsuji, H. Harada, N. Hariyama, L.
vegetative cell separation enzymes through a direct interaction with specific inhibitor
IseA in Bacillus subtilis. Mol. Microbiol. 70:168-182.
Figure legends
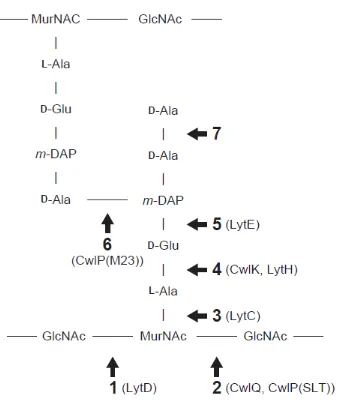
Fig. 1. Peptidoglycan structure and cleavage sites of autolysins. Peptidoglycan
structure of B. subtilis (GluNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; L-Ala, L-alanine; D-Glu, D-glutamic acid; m-DAP, meso-diaminopimelic acid;
D-Ala, D-alanine). Arrows with number indicate cleavage sites by autolysisns (1,
N-acetylglucosaminidase; 2, N-acetylmuraminidase and lytic transglucosidase; 3,
N-acetylmuramoyl-L-alanine amidase; 4, L,D-endopeptidase; 5, D,L-endopeptidase; 6,
D,D-endopeptidase; 7, D,D-carboxypeptidase). Catalytic domains of autolysisns which
are indicated in parentheses below the numbers, were used for chimeric enzyme in this study (Smith 2000).
Fig. 2. Expression and activity of the chimeric enzymes. Strains were cultured with
1% xylose for Pxyl-cwlO induction. Lanes; 1; , (A) Expression of the chimeric enzymes were evaluated by Western blot analysis with an anti-FLAG antibody. Degraded
products of the chimeric enzymes appeared in lane XXX and XXX. (B) Zymography of the chimeric enzymes using B. subtilis cell wall as a substrate. Arrows indicate clear zones produced by the chimeric enzymes.
Fig. 3. Localization of the chimeric enzymes. Strains were cultured with 1% xylose to
induce CwlO. Subcellular localization of the chimeric enzymes were observed by IFM using anti-FLAG antibody and anti-mouse IgG Cy3 conjugate antibody. A; , B; , ...
Fig. 4. Suppression of the lytE cwlO synthetic lethality by the chimeric enzymes.
TABLE 1. Bacterial strains and plasmids used in this study.
________________________________________________________________________________________________________
Name Relevant genotype Source or reference a
________________________________________________________________________________________________________
E. coli strains
JM109 recA1 endA1 gyrA96 thi-1 hsdR17 relA1 supE44 Δ (lac-proAB)
/F’ [traD36 proAB lacIq lacZ ΔM15] Takara C600 supE44 hsdR17 thi-1 thr-1 IeuB6 lacY1 tonA21 Laboratory stock M15/pREP4 lac ara gal mtl F- recA+ uvr+ / lacIq kan Qiagen
B. subtilis strains
168 trpC2 S. D. Ehrlich
OH001 trpC2 cwlO::pXyl-cwlO (Pxyl-cwlO) Hashimoto 2012
OH004 trpC2 lytE::lytE-6xFLAG cwlO::pXyl-cwlO (Pxyl-cwlO) Hashimoto 2012
XXX trpC2 lytE::pCA-NeClytD3 cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeClytD3 -> OH001
XXX trpC2 lytE::pCA-NeClytD4 cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeClytD4 -> OH001
MH02 trpC2 lytE::pCA-NeCcwlQ cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlQ -> OH001
MH03 trpC2 lytE::pCA-NeCcwlP(SLT) cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlP(SLT)
XXX -> OH001
XXX LytC
MH05 trpC2 lytE::pCA-NeCcwlK cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlK -> OH001
MH06 trpC2 lytE::pCA-NeClytH cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeClytH -> OH001
MH07 trpC2 lytE::pCA-NeCcwlP(M23) cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlP(M23)
-> OH001 XXX trpC2 lytE::pCA-NeCcwlP(M23)1580 cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlP(M23)H1580N
-> OH001 XXX trpC2 lytE::pCA-NeCcwlP(M23)1584 cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlP(M23)D1584Q
-> OH001 XXX trpC2 lytE::pCA-NeCcwlP(M23)1620 cwlO::pXyl-cwlO (Pxyl-cwlO) pCA-NeCcwlP(M23)H1620N
-> OH001
Plasmids
pCA-NeClytD3 bla cat NLytECLytD3 this study
pCA-NeClytD4 bla cat NLytECLytD4 this study
pCA-NeCcwlQ bla cat NLytECCwlQ this study
pCA-NeCcwlP(SLT) bla cat NLytECCwlP(SLT) this study
XXX LytC
pCA-NeCcwlK bla cat NLytECCwlK this study
pCA-NeClytH bla cat NLytECLytH this study
pCA-NeCcwlP(M23) bla cat NLytECCwlP(M23) this study pCA-NeCcwlP(M23)1580 bla cat NLytECCwlP(M23)H1580N this study pCA-NeCcwlP(M23)1584 bla cat NLytECCwlP(M23)D1584Q this study pCA-NeCcwlP(M23)1620 bla cat NLytECCwlP(M23)H1620N this study pHistkCwlF bla 6xHis-lytE (catalytic domain) Yamamoto 2008
pQE-M23 bla 6xHis-cwlP (M23 domain) Sudiarta 2010
________________________________________________________________________________________________________
a
Table S1. Primers used in this study.
________________________________________________________________________________________________________
CwlK-1 5’-tcttcaagcagcaaaacgtcatctacatcatgttttagttatattcggca -3’ CwlK-2 5’-gccggatccgttaggaatcatctccaag -3’
LytC-1 5’-tcttcaagcagcaaaacgtcatctacatcaaacactcctgccgtaagcac -3’ LytC-2 5’-gccggatcctctgtaataagatactgtgc -3’
LytD-2 5’-gcctctagatcgatattccggaacatcaa -3’
LytD-3 5’-tcttcaagcagcaaaacgtcatctacatcatcaacgaagctaaaacaatt -3’ LytD-4 5’-tcttcaagcagcaaaacgtcatctacatcaaagctaaaacaattatacgg -3’ LytE-19 5’-gcgcggtaccagctacgacagcagttg -3’
LytE-20 5’-tgatgtagatgacgttttg -3’
LytH-1 5’-tcttcaagcagcaaaacgtcatctacatcatctgaattcatgaagctgtt -3’ LytH-2 5’-gccggatcccttctttttttggtattcat -3’
YjbJ-1 5’-tcttcaagcagcaaaacgtcatctacatcagcagaaagcactgaaacag -3’ YjbJ-2 5’-gccggatcccgcgtaataaactgaagtta -3’
YomI-1 5’-tcttcaagcagcaaaacgtcatctacatcaggcaagtattcaagctaca -3’ YomI-2 5’-gccggatccggccatgatcttcttgacgt -3’
YomI-3 5’-cttcatcatcgtcttcatccggttcttcaaagaagatcatggccaact -3’ YomI-4 5’- gccggatcctttagcctgagctatccc -3’