The 3
rdNatural Pigments Conference For South East Asia
NP-SEA 2016
THE ANTIOXIDANT ACTIVITY OF CAROTENOID
PIGMENTS IN THE BACTERIAL SYMBIONTS OF
SEAGRASS
Syringodium isoetifolium
Delianis Pringgeniesa* and Riyanda Idrisa*
*aDepartment of Marine Sciences, Faculty of Fisheries and Marine Scinces, Diponegoro University, Semarang, Indonesia Abstract
Carotenoids are pigments of red, yellow and orange which are found in plants, animals and bacteria, and are known to have antioxidant activity. This study aims to identify the carotenoid pigments detected in seagrass Syringodium isoetifolium
bacterial symbionts. Isolation of bacteria was conducted using dispersive media Zobell 2116E. Bacterial isolates were cultured and then centrifuged at 8000 rpm for 10 minutes and extracted using methanol. Identification of the pigment was done by using High Performance Liquid Chromatography (HPLC) reversed phase ODS / C18. The mobile phase was carried out using a mixture of methanol: acetonitrile (7: 3 v / v). Free radical reduction activities determined by the method of DPPH (diphenylpicrylhydrazil) and its absorbance was measured at a wavelength of 517 nm. Identification of the bacterial symbionts from the seagrass S. isoetifolium performed using 16S rDNA PCR method. The results showed that, of the 12 bacterial isolates obtained, isolate 7A was proven to contain caratenoid pigment. Pigment extracts of the bacterial isolates had free radical DPPH reduction activity of 40.4%. The results showed that the identification of bacteria isolates 7A had 100% level of kinship with the bacteria Bacillus amyloliquifaciens.
Keywords: antioxidant activity, seagrass, pigment carotenoid, Bacillus amyloliquifaciens .*Corresponding author: Email address ([email protected])
Telephone Number: 081390800800
1. Introduction
Carotenoids are a class of biological pigments mostly consisting of the colors red, yellow and orange. It is one of the pigments with significant potential in healthcare applications. Carotenoids are not inherently manufactured within the biological system of humans and animals, and as such carotenoid intakes are taken from food. Food rich in carotenoids often come from land animals and plant. However, information on carotenoid-rich marine food sources is still scarce. One of such information was written by Arlita et al., (2013) who reported success in isolating bacteria Paracocusalcaliphilus
and Brevibacteriummaris from Caulerpa
cupresoides seaweed, which were attributed in producing carotenoids Xanthophyll and carotene. Moreover, Radjasa (2003) states that symbiont bacteria produce pigment similar to that of its host. Based on the findings above, symbiont bacteria can be a potential new source for carotenoids because these bacteria are environmentally friendly and can be mass-cultured in a relatively short amount of time. Carotenoid-producing symbiont bacteria can also be found in
Syringodium isoetifolium seagrass.
colors. This research aims to identify carotenoid pigments from symbiont bacteria of seagrass from the species
Syringodium isoetifolium with potential as a source for antioxidants.
2. Material and Methods
a. Sample Collection and Isolation of
Symbiont Bacteria
Samples of Syringodium isoetifolium
seagrass were collected the waters of Teluk Awur, Jepara, Indonesia. Seagrass samples weighing 5 grams were directly placed within sample tube. The tube was prepared prior to sample collation by adding sea water and storage in cool box. The samples were then cleaned off of surface bacteria with sterile sea water. The samples were then digested, and were mixed into 5 ml of sea water. This process yielded 100 of sample dilution. Of the Diluted sample, 0,5 ml was taken off and was transferred into a reaction tube with a sterile pipette. The reaction tube were prepared before by adding 4,5 ml of sterile sea water. This process yielded 10-1 of sample dilution. The processes were repeated until sample dilutions of 10-2, 10
-3, 10-4, 10-5, 10-6, and 10-7 were obtained.
Of each sample dilution factor, 100 μL of sample was dispersed over Zobell 2216E
media by using a spreader and all of the samples were incubated for 3 days in 30°C of temperature (Radjasa, 2003). Colonies displaying hues of yellow and orange (5.A.4) were selected and purified.
b. Identification of Symbiont Bacteria
DNA obtained from the 24-hours bacterial cultures were extracted using
High Pure PCR Template Preparation Kit (Roche). PCR 16S rDNA amplification was carried out by denaturation at 94°C for 5 minutes as initial heating, followed by 30 cycles (annealing at 94°C for 30 seconds, extension at 54°C for 60 seconds and rerun of denaturation process at 72°C for 120 seconds) and incubation at 4°C. The primer used in PCR 16S rDNA process is universal primer 27F (5'-AGAGTTTGATCMTGGCTCAG-3') and eubacteria-specific primer 1492R (5' TACGGYTACCTTGTTACGACTT-3') (Isnansetyo and Kamei, 2003). The electrophoresis process utilized agarose gel with 1 % concentration. The device was operated at 100 V for ± 45 minutes. The result obtained from electrophoresis process was observed under a UV Illuminator. The sequencing process was carried out in compliance with PCR
sequencing cycle using Big Dye
Terminator v.3.1 and resulting DNA sequences were compared with sequences in DNA database at Basic Local Alignment Search Tool (BLAST) of National Center
for Biotechnology Information, National Institute for Health, USA (www.ncbi.nlm.nih.gov) (Altschul et al., 1997).
c. Bacterial Culture and Extraction
until pellet was obtained, after which the pellet is weighed. The obtained pellets were diluted in methanol to separate between bacteria and its pigments, and were fixated by Nitrogen gas to obtain raw extracts. The obtained bacterial raw extracts were then dry-weighed before being put into another dilution process using methanol.
d. Pigment Identification
Raw extracts obtained from bacterial extraction were analyzed using UV-Vis CARY 50 at 190-800 nm wavelength setting and by using High-Performance
Liquid Chromatography (HPLC)
Shimadzu LC 20-AB reversed phase ODS/C18, 5 µl, 4 mm x 25 mm diameter with metanol:asetonitril (7:3 v/v) mobile phase, with flow rate of 1 ml. minute-1 and 1000 psi of pressure (Maeda, 2005). The analysis were then carried out 190-800 nm wavelength. HPLC results were processed and analyzed using OriginPro 8.1 software.
e. DPPH Test
DPPH test for bacterial pigment samples employed Molyneux (2004) method with several modifications. This method was performed by preparing 3 ml of DPPH stock 0,1 mM and added into 1 ml of test pigment solution (2000 ppm concentration) (control extract was replaced by methanol), incubated for 30 minutes in a dark room. Absorbance rate
was then calculated by using
spectrophotometer set at maximum
wavelength for DPPH (517 nm). inhibition rate calculation was also performed on the processed pigment sample.
3. Results and Discussion Identification of Bacteria
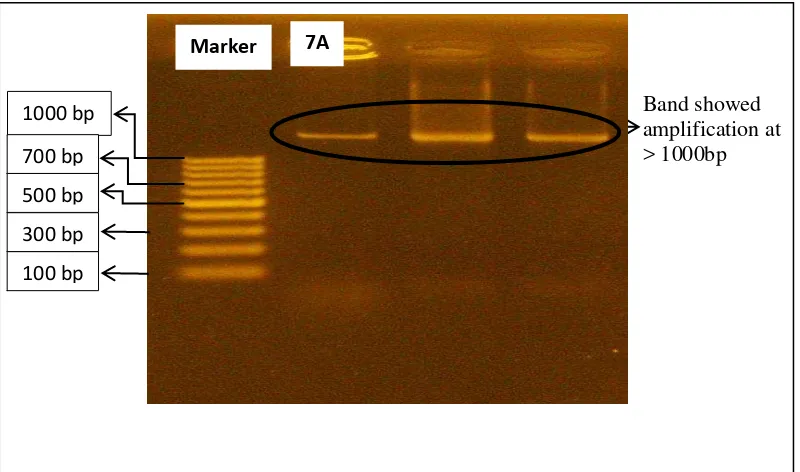
DNA amplification with PCR 16S rDNA showed positive results with DNA files displayed from the bacteria at the length of >1000 bp (Image 1). Primer used in this PCR 16S rDNA amplification was universal primer 27F and eubacteria-specific primer 1492R. Sequencing result in the form of base sequence of isolate 7A (>1000 bp) is displayed in Image 2.
Information: A= Adenine; T= Thymine; G= Guanine; C= Cytosine
Fig 2. Base Sequence Results of PCR 16S rDNA with Primer 27F on Isolate 7A Fig 1. Visualization of Resulting Band from PCR 16S rDNA on Sample Isolate
7A
1000 bp 700 bp 500 bp 300 bp 100 bp
7A Marker
Band showed amplification at > 1000bp
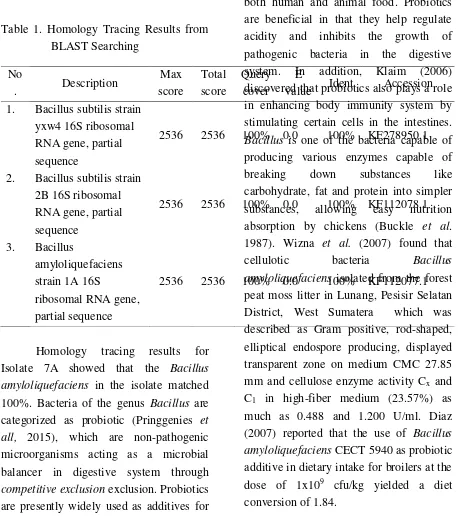
Table 1. Homology Tracing Results from
value Ident Accession
1. Bacillus subtilis strain yxw4 16S ribosomal RNA gene, partial sequence
2536 2536 100% 0.0 100% KF278950.1
2. Bacillus subtilis strain 2B 16S ribosomal
Homology tracing results for Isolate 7A showed that the Bacillus amyloliquefaciens in the isolate matched 100%. Bacteria of the genus Bacillus are categorized as probiotic (Pringgenies et all, 2015), which are non-pathogenic microorganisms acting as a microbial balancer in digestive system through
competitive exclusion exclusion. Probiotics are presently widely used as additives for
both human and animal food. Probiotics are beneficial in that they help regulate acidity and inhibits the growth of pathogenic bacteria in the digestive system. In addition, Klaim (2006) discovered that probiotics also plays a role in enhancing body immunity system by stimulating certain cells in the intestines.
Bacillus is one of the bacteria capable of producing various enzymes capable of
breaking down substances like
carbohydrate, fat and protein into simpler substances, allowing easy nutrition absorption by chickens (Buckle et al. 1987). Wizna et al. (2007) found that
cellulotic bacteria Bacillus
amyloliquefaciens isolated from the forest peat moss litter in Lunang, Pesisir Selatan District, West Sumatera which was described as Gram positive, rod-shaped, elliptical endospore producing, displayed transparent zone on medium CMC 27.85 mm and cellulose enzyme activity Cx and
C1 in high-fiber medium (23.57%) as
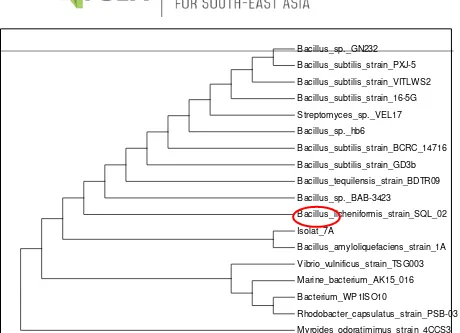
Fig 3. Phylogenetic Tree of Sequencing Results of 16S rDNA Isolate 7A
Identification of Pigment
Analysis result of Syringodium
duplicatum (7A) seagrass raw extract
using HPLC is displayed in Fig 4.
Spectrum pattern resulting from pigment
analysis on coral symbiont bacteria (7A)
can be seen in Image 5. Maximum
absorbance of HPLC analysis on
components of the extracts of coral
symbiont bacteria (7A) is presented in
Table 2.
Bacillus_sp._GN232
Bacillus_subtilis_strain_PXJ-5
Bacillus_subtilis_strain_VITLWS2
Bacillus_subtilis_strain_16-5G
Streptomyces_sp._VEL17
Bacillus_sp._hb6
Bacillus_subtilis_strain_BCRC_14716
Bacillus_subtilis_strain_GD3b
Bacillus_tequilensis_strain_BDTR09
Bacillus_sp._BAB-3423
Bacillus_licheniformis_strain_SQL_02
Isolat_7A
Bacillus_amyloliquefaciens_strain_1A
Vibrio_vulnificus_strain_TSG003
Marine_bacterium_AK15_016
Bacterium_WP1ISO10
Rhodobacter_capsulatus_strain_PSB-03
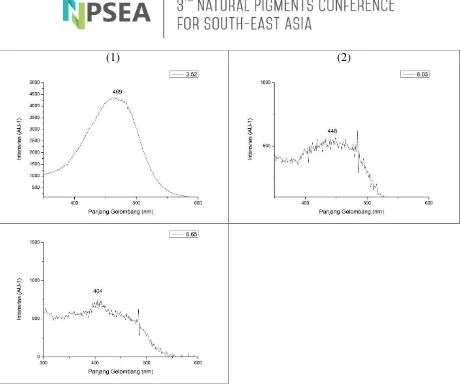
Fig 4. Chromatogram Profile of Raw Extract of Coral Symbiont Bacteria (7A) Displayed 3
Dominant Peaks.
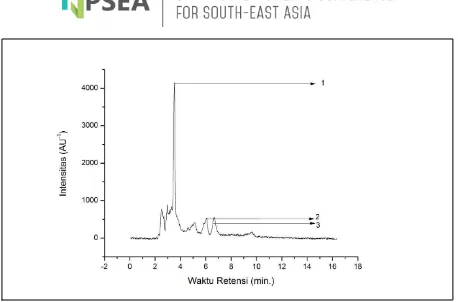
HPLC analysis of raw extract of
Syringodium isoetifolium seagrass symbiont bacteria (7A) resulted in 3 distinct dominant peaks with differing retention times. Detected peaks were
(1) (2)
Fig 5. Spectrum Pattern from Pigment Analysis on Seagrass Symbiont Bacteria (7.A) with 3.52 minute Retention Time (a), 6.03 minute Retention Time (b) and 6.65 minute Retention Time (c).
Table 2. Maximum Absorbance from HPLC of Seagrass Symbiont Bacteria Raw Extract Component (10.B.2)
Peak No. Retention
Time (Minutes)
Components Maximum Absorbance (nm)
Results Reference
1 3.52 Peridinin 469 473*
2 6.03 Fucoxanthin 448 449*
3 6.05 Unknown 404 -
* Based on Zapata et al. (2000)
Data processing on Origin 8.1 software resulted in different spectrum patterns for each chromatogram peaks. Carotenoid pigment was identified based
on spectrum pattern of each peak and its polarized order in comparison with results in a research by Zapata, et al.
(2000) which utilized relatively similar solution composition and static phase in
maximum absorbance rate of 404 nm which cannot be identified for pigments. This is due to the fact that the
corresponding retention time and
maximum absorbance rate of the peak did not match any of the category in the reference data. However, it is believed that the peak may correspond to a pigment belonging to xanthophyll class. Peak 1 with 3.52 minutes of retention time, and
maximum absorbance at 469 nm
wavelength highly matched the pigment Peridinin. Peak 2 with 6.03 minutes of retention time, and maximum absorbance at 448 nm wavelength was found to be the pigment Fucoxanthin. Fucoxanthin is a member of carotenoid pigment xantophyll and is often found as primary pigment in many species of brown algae. Fucoxanthin act as an active biological component, of which one of the benefit is anti-obesity. Fucoxanthin has shown to possess the capability stimulate the liver to produce DHA, although the mechanism of this bio-activity has not yet been fully understood. Fucoxanthin has been shown to be potential in its application to treat obesity as a new source of safer nTURl supplement (Nurcahyanti and Timotius, 2007). Fucoxanthin (Fx) also displayed antioxidant and other beneficial properties,
making it a very potential food supplement (Yasumoto, 2011).
DPPH Test
DPPH test was performed and analyzed using UV-Vis spectrophotometer at 517 nm wavelength. Any activity of free radical inhibition activity were observed from the color shift of DPPH solution after contact with sample and based on the maximum absorbance rate at 517 nm wavelength. Color shift was detected in DPPH solution in methanol after sample contact, which turned from violet to yellow (Fig 5). Spectrophotometer analysis of pigment raw extract from isolate 7A after 30 minutes of contact with DPPH is presented in Fig 5. Control maximum absorbance rate was recorded at 0.1884, where average maximum absorbance rate from 2 repetitions showed 0.112. This means that there was a 40.4% free radical inhibition from DPPH.
4. Conclusion
The research found that the symbiont bacteria of Syringodium isoetifolium
seagrass was Bacillus amyloliquefaciens. The bacteria produced Fucoxanthin, which showed capacity in inhibiting free radical from DPPH as much as 6.12%.
References
Altscul, S.F., T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller Arlita, N. L., Ocky K. R., Adi S. 2013.
Identifikasi Pigmen Karotenoid pada Bakteri Simbion Rumput Laut Caulerpa cupresoides (Vahl) C. Agardh. Journal of Marine Science. 2(3): Hal. 68-77.
Diaz-Ropero MP, Martin R, Sierra S, Lara-Villoslada F, Rodriguez JM, Xaus J, Olivares M. Two
Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J Appl Microbiol. 2007;102:337– 343. [PubMed]Diaz (2007)
Isnansetyo, A and Y. Kamei. 2003.
Anti-Methicillin-Resistant
Staphylococcus aureus
Substances. International Journal of Systematic and Evo. Microbiol. 53: Hal. 583-588.
Radjasa, O. K., 2003. Marine Invertebrate-Associated Bakteria in Coral Reef Ecosystems as a New Source of Bioaktive Compounds. J. Coast. Dev. 7: Hal. 65-70.
Klaim. 2006. The Online Encyclopaedia. Wikipedia. probiotik juga ikut berperan dalam meningkatkan kekebalan tubuh
Maeda, H., M. Hosokawa, T. Sashima, K.
Funayama dan K.
Miyashita.2005. Fucoxanthin
From Edible Seaweed, Undaria pinnatifida, Shows Antiobesity Effect Through UCP1 Expression
in White Adipose Tissues.
Biochemical and Biophysical Research Communications. 332: Hal. 392-397.
Molyneux. P. 2004. The Use of the Stable
Free Radical
Diphenylpicrilhidrazyl (DPPH)
for Estimating Antioxidant
Activity. Songklanarin J. Sci. Technol. 26: Hal. 211-219.
Nugraheni, S. A., M. M. Khoeri, L. Kusmita, Y. Widyastuti dan O. K. Radjasa. 2010. Characterization of Carotenoid Pigments from
Bacterial Symbionts of Seagrass
Thalassia hemprichii. J. Coast. Dev. 14(1): Hal. 51-60
Pringgenies. D., Izzuddin Azmi, Ali Ridho, Riyanda Idris. 2015
Exploration of Bacteria
Symbionts Mangrove Waste For The Production of Decomposter. Proceeding on “International
Conference on Coastal Zone”
Osaka, Japan May 16-18, 2016 June 2015.
Wizna, H. Abbas, Y. Rizal, A. Dharma & I. P. Kompiang. 2007. Selection and identification of cellulase-producing bacteria isolated from the litter of mountain and swampy forest. J. Microbiology Indonesia, 1(3):135-139.
Yamamoto K, Ishikawa C, Katano H, Yasumoto T, Mori N. 2011. Fucoxanthin and its deacetylated product, fucoxanthinol, induce apoptosis of primary effusion lymphomas. Cancer Lett ;300:225–234. [PubMed
Zapata M., Francisco R. dan Jose L. G. 2000. Separation of Chlorophtlls and Carotenoids from Marine Phytoplankton: A New HPLC Methode Using a Reversed Phase
C8 Column and