www.elsevier.com/locate/ibmb
Identification of a point mutation in an esterase gene in different
populations of the southern cattle tick, Boophilus microplus
Ruben Hernandez
a, b, Haiqi He
c, Andrew C. Chen
c, Suryakant D. Waghela
a,
G. Wayne Ivie
c, John E. George
d, G. Gale Wagner
a,*aDepartment of Veterinary Pathobiology, College of Veterinary Medicine, Texas A&M University, College Station, TX. 77843-4467, USA bCenid-Parasitologia Veterinaria INIFAP-SAGAR, Apartado Postal 206 Civac, Morelos, CP 62500, Mexico
cFood Animal Protection Research Laboratory, USDA-ARS, 2881 F&B Road, College Station, TX 77845, USA
dKnipling-Bushland US Livestock Insects Research Laboratory, USDA-ARS, 2700 Fredericksburg Road, Kerrville, TX 78028, USA
Received 13 October 1999; received in revised form 28 February 2000; accepted 6 March 2000
Abstract
Two esterase cDNA sequences were obtained from susceptible and organophosphorus resistant strains of Boophilus microplus. Both sequences have a high degree of homology to carboxylesterase B. One gene has identical sequences in both strains and the other showed two point mutations. One mutation produces an amino acid substitution when the amino acid sequence is deduced, this mutation was detected in six different populations susceptible and resistant to insecticides, but a pyrethroid resistant strain was the only one that showed only the mutant allele. Identification of this mutation and the strong signal detected in southern blot with this strain, suggest that esterases are contributing to detoxification of pyrethroid compounds, as a resistant mechanism in Mexican strains of the southern cattle tick. 2000 Elsevier Science Ltd. All rights reserved.
Keywords: Boophilus microplus; Esterases; Point mutation; Pyrethroid resistance; Southern blot
1. Introduction
The southern cattle tick, Boophilus microplus, is present in tropical and subtropical regions throughout the world. This tick species is detrimental to the cattle indus-try, not only because of its direct effect on production, but also its ability to transmit highly virulent hemoparas-ites such as Babesia spp. and Anaplasma marginale. In recent years the control of this tick has become difficult because of the development of resistance to the acari-cides used for its control (Nolan and Schnitzerling, 1986).
In many arthropod pests, resistance to pesticides has been attributed to an increase in pesticide degrading enzyme activity. However, in most cases it is not known whether this arises from mutant enzymes or from the
* Corresponding author. Tel.:+1-409-845-4275; fax: +1-409-862-2344.
E-mail address: [email protected] (G. Gale Wagner).
0965-1748/00/$ - see front matter2000 Elsevier Science Ltd. All rights reserved. PII: S 0 9 6 5 - 1 7 4 8 ( 0 0 ) 0 0 0 6 9 - 2
increased production of an enzyme already present in susceptible insects (Field et al., 1988).
Most insecticides in use today are esters of substituted phosphoric, carbamic or cyclopropanecarboxilic acids, and are consequently subject to degradation by esterases (Devonshire, 1991). The massive overproduction of any esterase by insects may result in the increased detox-ification of insecticide esters first by sequestration and then by hydrolysis when the inhibited esterase reactiv-ates (Devonshire and Field, 1991). Increased hydrolysis of insecticides by esterases has been implicated in insec-ticide resistance in many species. Recently, a carboxyle-sterase with a single amino acid substitution was shown to contribute significantly to organophosphorus (OP) resistance in blowflies (Newcomb et al., 1997). Also in both the mosquito Culex quinquefasciatus (Mouches et al., 1986) and the aphid Myzus persicae (Field et al., 1988), amplification of esterase genes is related with increased detoxification of insecticides. Rosario-Cruz et al. (1997) found that an OP resistant strain of B.
microplus contained several different proteins showing
activity when compared with a susceptible strain. Simi-larly, pyrethroid-hydrolyzing esterases have also been detected in B. microplus (De Jersey et al., 1985).
In this study we report the complete cDNA sequences of two esterases from B. microplus, a point mutation in one sequence, and the differences among susceptible and resistant strains in Southern hybridization.
2. Materials and methods
2.1. Ticks
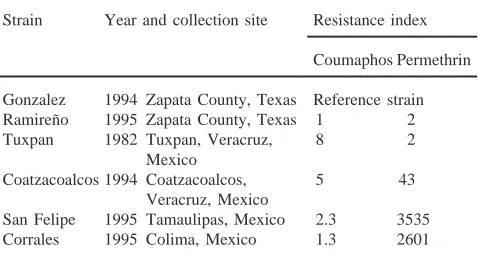
Unfed larvae of six B. microplus strains originally obtained from various locations in Texas and Mexico, and maintained at the Cattle Fever Tick Research Lab-oratory, ARS, USDA, at Mission, TX, USA were used in this investigation (Table 1). The Gonzalez and Ramiren˜o strains were obtained from two separate outbreaks of ticks in Texas. Both strains are susceptible to OP and pyrethroid compounds, and have been maintained with-out any insecticidal pressure. The Tuxpan and Coatza-coalcos strains resistant to OP and pyrethroid compounds respectively, were originally obtained from Veracruz in Mexico. Coatzacoalcos strain also exhibits a moderate level of OP resistance of 5 fold to coumaphos. The San Felipe and Corrales strains were obtained from Tamaul-ipas and Colima, Mexico respectively, both showing high resistance to permethrin. Assessment of suscepti-bility to acaricides was performed using the larval packet test (Stone and Haydock, 1962), with the Gonzalez strain as the susceptible reference. The resistant strains have been maintained under constant chemical pressure since their arrival at the laboratory in Mission, Texas. Each generation of Tuxpan strain has been exposed to increas-ing concentrations of coumaphos, at a rate of 0.05% of active ingredient. Coatzacoalcos, San Felipe, and Cor-rales strains have been similarly exposed to permethrin.
Table 1
B. microplus strains and resistance levels to coumaphos (OP) and
per-methrin (Pyrethroid)
Strain Year and collection site Resistance index
Coumaphos Permethrin
Gonzalez 1994 Zapata County, Texas Reference strain Ramiren˜o 1995 Zapata County, Texas 1 2 Tuxpan 1982 Tuxpan, Veracruz, 8 2
Mexico
Coatzacoalcos 1994 Coatzacoalcos, 5 43 Veracruz, Mexico
San Felipe 1995 Tamaulipas, Mexico 2.3 3535 Corrales 1995 Colima, Mexico 1.3 2601
2.2. DNA isolation
Total genomic DNA was obtained from tick larvae by phenol extraction using a modification of the method described by Sambrook et al., (1989). Briefly, 1 g of larvae was pulverized in liquid nitrogen with a mortar and pestle. After the liquid nitrogen had dissipated the sample was mixed with 10 ml of extraction buffer (Tris, 10 mM; EDTA, 100 mM; SDS, 0.5%; pH 8.0), the DNA was extracted with phenol/chloroform and then precipi-tated with ethanol in the presence of 300 mM NaCl. The DNA pellet was dissolved in the extraction buffer and
centrifuged at 110 000g for 20 min at 4°C to remove
glycogen. The DNA solution was then treated success-ively with RNase A, proteinase K, phenol extracted, and ethanol precipitated. The final DNA preparation had an A260/A280 .1.8.
2.3. RNA isolation and cDNA synthesis
Total RNA was isolated from tick larvae using the TRIzol reagent (Life Technologies, Gaithersburg, MD). 1 g of larvae was pulverized in liquid nitrogen as described above, then it was homogenized on ice in 10 ml of TRIzol reagent using a glass grinder with a Teflon pestle. Samples were incubated at room temperature for 5 min and centrifuged at 272g to remove cuticle and tissue particles. The remaining purification steps
fol-lowed the manufacturer’s protocol. Poly A+mRNA was
purified from the total RNA using the Oligotex mRNA purification kit (Qiagen, Santa Clarita, CA). cDNA syn-thesis and rapid amplification of cDNA ends (RACE) were performed using the Marathon cDNA Amplifi-cation kit (Clontech, Palo Alto, CA) following the manu-facturer’s protocol.
2.4. Oligonucleotide primers
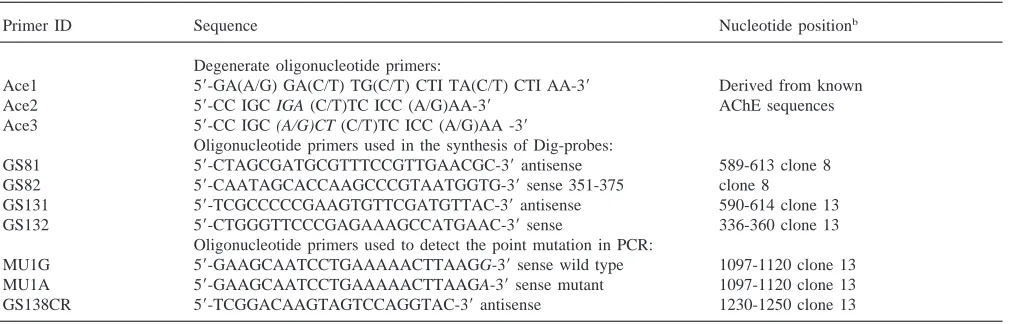
Degenerate oligonucleotide primers Ace1, Ace2, and Ace3 were chosen based on sequences of amino acids conserved in acetylcholinesterase (AChE) from other organisms, following the previously described methods (Arpagaus et al., 1994). Also specific primers were derived from clones 8 and 13 and used in the synthesis of probes labeled with digoxigenin, and in detection of a point mutation (Table 2).
2.5. Polymerase Chain Reaction (PCR)
PCR was carried out in a GeneAmp 9600 thermal cycler (Perkin Elmer Cetus, Norwalk, CT). The reaction mixture to obtain the first fragment with the degenerate
primers contained the 4 deoxyribonucleoside 59
-triphos-phates (200 µM each), degenerate primers Ace1 and
Ace2 or Ace3 (2 µM each), adaptor-ligated cDNA as
Table 2
Primers used in the PCR reactionsa
Primer ID Sequence Nucleotide positionb
Degenerate oligonucleotide primers:
Ace1 59-GA(A/G) GA(C/T) TG(C/T) CTI TA(C/T) CTI AA-39 Derived from known
Ace2 59-CC IGC IGA (C/T)TC ICC (A/G)AA-39 AChE sequences
Ace3 59-CC IGC (A/G)CT (C/T)TC ICC (A/G)AA -39
Oligonucleotide primers used in the synthesis of Dig-probes:
GS81 59-CTAGCGATGCGTTTCCGTTGAACGC-39antisense 589-613 clone 8 GS82 59-CAATAGCACCAAGCCCGTAATGGTG-39sense 351-375 clone 8
GS131 59-TCGCCCCCGAAGTGTTCGATGTTAC-39antisense 590-614 clone 13
GS132 59-CTGGGTTCCCGAGAAAGCCATGAAC-39sense 336-360 clone 13
Oligonucleotide primers used to detect the point mutation in PCR:
MU1G 59-GAAGCAATCCTGAAAAACTTAAGG-39sense wild type 1097-1120 clone 13 MU1A 59-GAAGCAATCCTGAAAAACTTAAGA-39sense mutant 1097-1120 clone 13 GS138CR 59-TCGGACAAGTAGTCCAGGTAC-39antisense 1230-1250 clone 13
aI=deoxyinosine. Ace1=sense primer derived from the amino acid sequence EDCLYLN conserved in the AChE family. Ace2 and Ace3= antisense primers derived from the amino acid sequence FGESAG conserved in the AChE family, differing in the codon for serine (TCI or AG(T/C) underlined. MU1G and MU1A different in the last nucleotide, underlined.
b Based on nucleotide sequences from clones 8 and 13, considering ATG start codon as nucleotide one.
(10 mM, pH 8.3), KCl (50 mM), MgCl2(1.5 mM), and
gelatin (0.001%) in a final volume of 25µl. The reaction mixture was initially heated for 3 min at 94°C, then
incu-bated for 40 cycles (94°C for 1 min, 46°C for 1 min,
and 72°C for 1.5 min), and a final extension for 5 min
at 72°C. All reagents for PCR were obtained from Perkin
Elmer, except primers, which were obtained from Inte-grated DNA Technologies Inc. (Coralville, IA). PCR products were separated by electrophoresis on 1% aga-rose gels (SeaKem ME, FMC, Rockland, ME) in TBE buffer (Tris 45 mM, borate 45 mM, EDTA 1 mM, pH 8.0). The resulting bands were visualized by ethidium bromide staining.
Reaction mixture for PCR to obtain specific fragments with the wild type or the mutant allele contained the
4 deoxyribonucleoside 59-triphosphates (200 µM each),
specific primers GS138CR and MU1G or MU1A (0.2
µM), genomic DNA as template, Advantage cDNA
polymerase mix (0.5 units), Tricine-KOH (40 mM),
KOAc (15 mM), Mg(OAc)2(3.5 mM), and bovine serum
albumin (3.75 µg/ml) in a final volume of 25µl.
Tem-perature conditions included 3 min at 94°C, then 35
cycles (94°C for 20 sec, 63°C for 30 sec, and 68°C for
30 sec), and a final extension for 5 min at 68°C. All
reagents for PCR were obtained from Clontech except primers, which were obtained from Integrated DNA Technologies Inc. (Coralville, IA). PCR products were separated and visualized as above.
2.6. cDNA cloning
PCR products were isolated and purified from gel slices using the Advantage PCR-Pure kit (Clontech). Purified products were end-polished using the PCR Pol-ishing kit (Stratagene, La Jolla, CA). The polished DNA
fragments were cloned using the Zero Blunt PCR clon-ing kit, and transformed into TOPO10 competent cells (Invitrogen, Carlsbad, CA). Colonies were screened by PCR with M13 universal primers for the presence of DNA inserts. Plasmid DNA was purified from positive colonies using a plasmid miniprep kit (BIO-RAD Labs, Hercules, CA).
2.7. cDNA sequencing
Recombinant plasmids were sequenced on an auto-mated DNA sequencer (ABI-PRISM 377, Perkin Elmer, Foster City, CA) using the dye-terminator at the DNA Sequencing and Oligo Lab, Department of Veterinary Pathobiology, Texas A&M University, College Station, TX. Sequences were analyzed using the BCM Search Launcher (Human Genome Center, Baylor College of Medicine, Houston, TX) on the Internet
(:8088/search-launcher/launcher.html). From plasmid inserts that
showed homology with published sequences of AChE,
we designed specific primers that were used for 59- and
39-RACE to obtain the entire cDNA sequence.
2.8. Southern hybridization
Genomic DNA digested with either restriction endon-uclease EcoRI or double digested with EcoRI and NsiI was separated in 1% agarose gel by electrophoresis stained with ethidium bromide and photographed. The DNA in the gel was denatured, neutralized and
trans-ferred to a Nylon membrane (Nytran, Schleicher and
Schuell, Keene, NH) as described by Sambrook et al.
(1989). The membrane was then rinsed with 2×SSC and
baked at 80°C for 2 h in a vacuum oven. Two probes
(Table 2) with the PCR DIG Probe Synthesis Kit (Boehringer Mannheim, Indianapolis, IN), and using Gonzalez cDNA as the template. PCR conditions
included denaturation at 95°C for 5 min, then 5 cycles
of 94°C for 30 s and 72°C for 4 min, 5 cycles of 94°C
for 30 s and 70°C for 4 min, 35 cycles of 94°C for 20
s and 68°C for 4 min, and a final extension of 68°C for 5 min. Hybridization and signal detection were carried out using the DIG High Prime DNA Labeling and Detec-tion Starter kit II (Boehringer-Mannheim). Briefly, the baked Nylon membrane was prehybridized with a stan-dard hybridization buffer containing formamide for 30
min at 48°C; the probe was denatured by boiling for 5
min, added to the prehybridization solution and
incu-bated in a hybridization oven for 12 h at 48°C with
gentle agitation. High stringency conditions included 2
post-hybridization washes of 5 min each in 2×SSC and
0.1% SDS at room temperature and 2 washes of 15 min
each in 0.5×SSC and 0.1% SDS at 68°C under constant
agitation. Finally the labeled DNA bands were detected following the manufacturer’s protocol, after exposing an X-ray film (BioMax MR film, Eastman Kodak Com-pany, Rochester, N.Y.) to the membrane for 18 h.
3. Results
A single DNA band of <300 bp obtained by using
primers Acel and Ace3 in PCR was isolated and cloned. Twenty clones were selected, seven of which were sequenced and three different sequences showing a 330 bp fragment were obtained. These three sequences were compared with published sequences in the GenBank showing similarity to known AChEs. We have reported one sequence that encodes a putative acetylcholinester-ase (Hernandez et al., 1999). By using gene specific
pri-mers obtained from the other two sequences in the 59
-and 39-RACE and subsequent primer walking, we
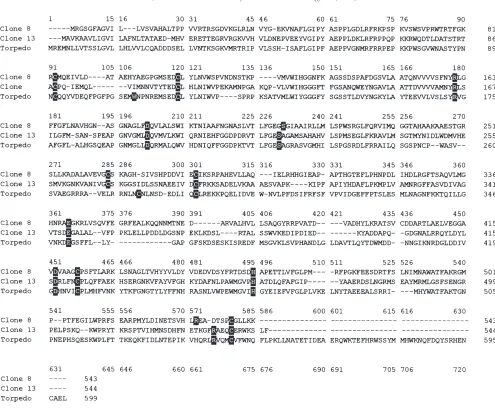
obtained the entire sequence of two esterase cDNAs. The cDNAs had an open reading frame of 1629 bp for clone 8 (Fig. 1) and 1632 bp for clone 13 (Fig. 2) encoding proteins of 543 and 544 amino acids respectively. Con-served amino acids in the cholinesterase family present in Torpedo marmorata (Bon et al., 1986) were also found at the corresponding positions in our sequences from B. microplus: Ser 224, His 464, Glu 351, 3 pairs of Cys residues (91-118, 278-289, and 426-545), Asp 196, Asp 421, Arg 173, and Arg 541 (Fig. 3). However, tryptophan 108 reported in T. marmorata as a critical component of the choline-binding site is absent in both sequences reported here.
Analysis of the deduced amino acid sequences from both clones using the program GenomeNet www.server on the Internet (), showed motifs for protein kinase and casein kinase phosphorylation sites, as well as N-myris-toylation and N-glycosylation sites. Motifs were present
indicating that both proteins are type-B carboxylester-ases. The hydropathy index analysis revealed that clone 13 codes for a membrane protein that has one transmem-brane helix in the N-terminal region. In contrast, clone 8 was identified as a soluble protein using the BCM Search Launcher: Protein Secondary Structure Prediction on the Internet (:9331/seq-search/struc-predict.html).
cDNA synthesized from the Tuxpan strain was sequenced using specific primers obtained for the zalez susceptible strain. Alignment of cDNA from Gon-zalez and Tuxpan strains showed identical nucleotide sequences in clone 8, but two point mutations were detected when sequences derived from clone 13 were compared. One is a silent mutation that does not change
the amino acid sequence (nucleotide 1458 T→C),
whereas the other (nucleotide 1120 G→A) changes an
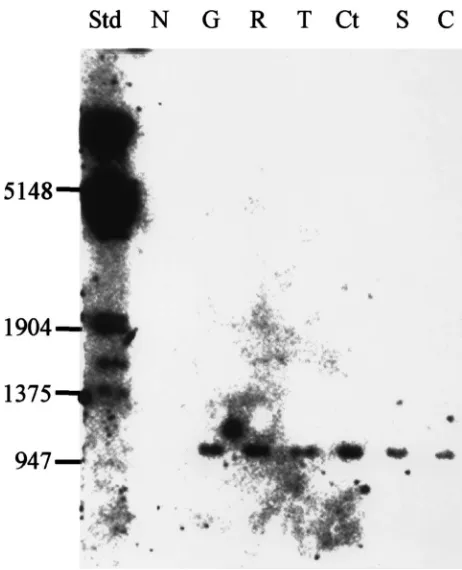
aspartate to an asparagine, substituting a polar uncharged amino acid for a negative charged (Figs. 2 and 3). When genomic DNA was amplified using the specific primer pairs GS138CR-MU1G or GS138CR-MU1A, which amplify wild type (G) or mutant (A), respectively, both alleles were found in all strains examined in this study except the Coatzacoalcos strain, which showed only the mutant allele (Fig. 4).
Two digoxigenin-labeled probes derived from clones 8 and 13 were used to analyze genomic DNA digested with restriction endonucleases. When restriction frag-ments of digestion with 2 nucleases (EcoRI and NsiI) were probed with a 263 bp oligonucleotide derived from clone 8, a single band of the same size was detected in all 6 strains (Fig. 5). When EcoRI digested DNA was probed with a 279 bp oligonucleotide derived from clone 13, two different restriction patterns were revealed. The Gonzalez, Ramiren˜o, Tuxpan, San Felipe and Corrales strains showed two fragments between 10 and 20 kb, whereas the Coatzacoalcos strain showed a single frag-ment <10 kb, with a stronger hybridization signal than
the other strains (Fig. 6). The strong signal was con-served in the Coatzacoalcos strain when hybridization was carried out with DNA digested with two restric-tion enzymes.
Genomic DNA from the six strains was amplified by PCR with the same primers used to produce probes 8 (GS81-GS82) and 13 (GS131-GS132). These genomic DNA fragments maintained the same size as the cDNA fragments indicating that there were no intronic sequences in these specific regions (data not shown).
4. Discussion
Fig. 3. Amino acid sequence alignment with electric ray AChE. Deduced amino acid sequences from cDNA clones 8 and 13 from B. microplus are aligned with AChE sequence of Torpedo marmorata. Conserved amino acids present in T. marmorata that play an important role in the catalytic activity and their corresponding amino acids in B. microplus are printed in white on a black background.
Fig. 4. Amplified products using different primers. PCR products were generated using primer GS138CR paired with primers with a single nucleotide substitution, MU1G or MU1A in the 39end. Genomic DNA from six different strains was used as templates. The products were analyzed on 1% agarose gel. G, Gonzalez; R, Ramiren˜o; T, Tux-pan; Ct, Coatzacoalcos; S, San Felipe; C, Corrales; Std, DNA standards (in bp); A, cDNA from Gonzalez.
they both contain a motif for the type B ase. In Drosophila melanogaster over 30 carboxylester-ases have been identified (Oakeshott et al., 1993), and it is probable that a large number of esterases is also present in B. microplus.
While both sequences reported here appear to encode carboxylesterase, substantial differences exist. A well-conserved sequence, GXSXG, used as a signature pat-tern in the esterase family is present in clone 13 (GESAG), but the corresponding segment in clone 8 is replaced by a different sequence (GEGSG). Results of motif finder on the Internet (), label the clone 8 as a carboxylesterase B2, and clone 13 as a carboxylesterase B1. Increased production of esterase B1 has been found to be responsible for resistance to OP insecticides in
Culex mosquitoes (Mouches et al., 1986). While clone
Fig. 5. Southern hybridization with probe 8. Genomic DNA from 6 different strains was digested with restriction enzymes EcoRI and NsiI.
5µg of digested DNA from each strain was separated on a 1% agarose
gel. After transferring to Nylon membrane the fragments were hybridized with a specific digoxigenin-labeled probe (263 bp) derived from the clone 8 cDNA. Std, DNA standards in bp; N, PCR product derived from a different esterase cDNA clone used as a negative con-trol; G, Gonzalez; R, Ramiren˜o; T, Tuxpan; Ct, Coatzacoalcos; S, San Felipe; C, Corrales.
The results of amplification of genomic DNA using primers that differ by only one base, show that all strains used in this investigation possess the mutant allele and that the Coatzacoalcos strain is the only one that lacks the wild type allele. Working with some of these same strains of ticks, Miller and coworkers (Miller et al., 1999) found that triphenylphosphate (TPP), an esterase inhibitor, increases the toxicity of pesticides in the Coat-zacoalcos strain, indicating that esterases are a major mechanism of resistance in this strain. In contrast, TPP did not have any effects on the Corrales and San Felipe strains. Jamroz et al. (in press) found significant enhancement of permethrin hydrolysis and increased general esterase activity in the Coatzacoalcos strain, with no enhancement of similar activity in either the Corrales or the San Felipe strains. They concluded that metabolic detoxification of pyrethroids by carboxylesterases is an important resistance mechanism in this strain, while tar-get site pyrethroid insensitivity seems to play an important role in Corrales and San Felipe strains. He et al. (1999) found a mutation in the sodium channel gene in San Felipe and Corrales strains but that mutation was absent in the Coatzacoalcos strain. These results indicate
Fig. 6. Southern hybridization with probe 13. Genomic DNA from 6 different strains was digested with restriction enzyme EcoRI. 5µg of digested DNA from each strain was separated on a 1% agarose gel. After transferring to Nylon membrane the fragments were hybridized with a specific digoxigenin-labeled probe (279 bp) derived from the clone 13 cDNA. Std, DNA standards in bp; N, PCR product derived from a different esterase cDNA clone used as a negative control; G, Gonzalez; R, Ramiren˜o; T, Tuxpan; Ct, Coatzacoalcos; S, San Felipe; C, Corrales.
that the mutated esterase gene may be involved in meta-bolic detoxification of pyrethroid compounds. Also they indicate that both target site modification and detoxify-ing esterases are involved in these Mexican pyrethroid-resistant strains.
Another possible way for esterases to be involved in resistance is mutated esterases that are refractory to pes-ticide treatment, and mutations that result in more efficient enzymes to hydrolyze or to increase seques-tering of pesticides. A mutation leading to a single amino acid substitution has been shown to convert a carboxyle-sterase to an enzyme that hydrolyzes OP compounds more efficiently in a blow fly (Newcomb et al., 1997).
It is possible that these mechanisms might be operating together in a tick to bring about resistance. Only further investigation can determine the relative contribution of each factor and to see if the mutant allele is related with a specific type of resistance or resistance to a specific family of insecticides. The presence of both alleles in the majority of the strains indicates that they are mixed populations; now it is important to know the frequency rate at which the mutant allele may be present. Research is currently underway to identify the mutation in genomic DNA from single tick larvae, and to find how the mutation is distributed within a population. The presence of this mutation at a higher frequency in resist-ant populations of ticks would have far reaching impli-cations in the management of tick control programs, and the use of insecticides more effectively.
Acknowledgements
We thank Ronald B. Davey for providing the ticks used in this study. Ruben Hernandez was sponsored by Consejo Nacional de Ciencia y Tecnologia, Mexico (54249). Mention of a commercial or proprietary product in this paper does not constitute an endorsement of this product by the USDA, nor does it imply the recommen-dation of the product by the USDA to the exclusion of similar products.
References
Arpagaus, M., Fedon, Y., Cousin, X., Chatonnet, A., Berge, J.-B., Fournier, D., Toutant, J.-P., 1994. cDNA sequence, gene structure, and in vitro expression of ace-1, the gene encoding acetylcholines-terase of class A in the nematode Caenorhabditis elegans. J. Biol. Chem. 269, 9957–9965.
Bon, S., Chang, J.Y., Strosberg, A.D., 1986. Identical N-terminal pep-tide sequences of asymmetric forms and of low-salt-soluble and detergent-soluble amphiphilic dimers of Torpedo
acetylcholinester-ase. Comparison with bovine acetylcholinesteracetylcholinester-ase. FEBS Letters. 209, 206–212.
De Jersey, J., Nolan, J., Davey, P.A., Riddles, P.W., 1985. Separation and characterization of the pyrethroid-hydrolyzing esterases of the cattle tick, Boophilus microplus. Pest. Biochem. Physiol. 23, 349–357.
Devonshire, A.L., 1991. Role of esterases in resistance of insects to insecticides. Biochem. Soc. Trans. 19, 755–759.
Devonshire, A.L., Field, L.M., 1991. Gene amplification and insecti-cide resistance. Annu. Rev. Entomol. 36, 1–23.
Field, L.M., Devonshire, A.L., Forde, B.G., 1988. Molecular evidence that insecticide resistance in peach-potato aphids (Myzus persicae Sulz.) results from amplification of an esterase gene. Biochem. J. 251, 309–312.
Guillemaud, T., Makate, N., Raymond, M., Hirst, B., Callaghan, A., 1997. Esterase gene amplification in Culex pipiens. Insect Mol. Biol. 6, 319–327.
He, H., Chen, A.C., Davey, R.D., Ivie, G.W., George, J.E., 1999. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle tick. Biochem. Biophys. Res. Commun. 261, 558–561.
Hernandez, R., He, H., Chen, A.C., Ivie, G.W., George, J.E., Wagner, G.G., 1999. Cloning and sequencing of a putative acetylcholinester-ase cDNA from Boophilus microplus (Acari: Ixodidae). J. Med. Entomol. 36, 764–770.
Jamroz, R.C., Guerrero, F.D., Pruett, J.H., Oehler, D.D., Miller, R.J., 2000. Molecular and biochemical survey of acaricide resistance mechanisms in larvae from Mexican strains of the southern cattle tick, Boophilus microplus. J. Insect Physiol. (in press).
Miller, R.J., Davey, R.B., George, J.E., 1999. Characterization of pyr-ethroid resistance and susceptibility to coumaphos in Mexican
Boophilus microplus (Acari: Ixodidae). J. Med. Entomol. 36,
533–538.
Mouches, C., Pasteur, N., Berge, J.B., Hyrien, O., Raymond, M., De Saint Vincent, B.R., De Silvestri, M., Georghiou, G.P., 1986. Amplification of an esterase gene is responsible for insecticide resistance in a California Culex mosquito. Science 233, 778–780. Newcomb, R.D., Campbell, P.M., Ollis, D.L., Cheah, E., Russell, R.J., Oakeshott, J.G., 1997. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. 94, 7464–7468.
Nolan, J., Schnitzerling, H.J., 1986. Drug resistance in arthropod para-sites. In: Campbell, W.C., Rew, R.S. (Eds.), Chemotherapy of Para-sitic Diseases. Plenum, New York and London, pp. 603–620. Oakeshott, J.G., van Papenrecht, E.A., Boyce, T.M., Healy, M.J.,
Rus-sell, R.J., 1993. Evolutionary genetics of Drosophila esterases. Genetica 90, 239–268.
Rosario-Cruz, R., Miranda-Miranda, E., Garcia-Vazquez, Z., Ortiz-Estrada, M., 1997. Detection of esterase activity in susceptible and organophosphate resistant strains of the cattle tick Boophilus
microplus (Acari: Ixodidae). Bull. Entomol. Res. 87, 197–202.
Sambrook, J., Fritsch, E.F., Maniatis, T., 1989. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.