The focus of research on signalling in Rhizobium–legume interactions has moved from understanding the structure and synthesis of rhizobially made Nod factors, towards an analysis of how they function in plants. Nod-factor-induced changes in ion fluxes across membranes, followed by establishment of an oscillation of intracellular Ca2+concentration, point to the involvement of a receptor-mediated signal transduction pathway. Progress towards the identification of components in this pathway is being made by identifying Nod-factor binding proteins, isolating plant mutants that are defective in signalling and analysing plant responses to Nod factors.
Addresses
John Innes Centre, Norwich Research Park, Colney, Norwich, NR4 7UH, UK
*e-mail: [email protected]
†e-mail: [email protected]
Current Opinion in Plant Biology1999, 2:483–489
1369-5266/99/$ — see front matter © 1999 Elsevier Science Ltd. All rights reserved.
Abbreviations
ENOD early nodulation
LCO lipo-chitin oligomer
LNP lectin nucleotide phosphohydrolase
NFBS Nod-factor-binding site
Introduction
Nod factors, which are signalling molecules made by rhi-zobia, initiate nodule development in legumes. They are composed of lipo-chitin oligomers (LCOs) that usually comprise four or five β, 1–4 linked N-acetyl glucosamine residues, in which the N-acetyl group of the terminal (non-reducing) sugar is replaced by an acyl chain. Many Nod factors, made by a diverse range of rhizobia, have been
characterised and it is evident that specificity (i.e. the types of legume hosts nodulated by given rhizobia) is determined, at least in part, by chemical modification of the Nod factors. These modifications include the attach-ment of sulphate, acetate, carbamoyl groups; addition of other sugars such as arabinose, mannose or fucose (and substituted derivatives of fucose); changes to the acyl chain; and variation of the chitin oligomer length (Figure 1). (The role of rhizobial nodgene products in Nod factor biosynthesis has been reviewed in detail [1,2,3•].)
In the appropriate legume, Nod factors can induce a vari-ety of effects including deformation of root hairs, division of root cortical cells, and nodule morphogenesis [1,4•,5••].
Some of these responses are induced at concentrations as low as 10–12–10–13M Nod factor; this finding alone points
to the probable existence of high-affinity receptors. Other components may act to enhance nodulation; thus, for example, a protein secreted by some rhizobia can extend the range of legumes nodulated [6,7] possibly by forming ion channels in the plant membranes [8]. Proteins secreted by other rhizobia are also likely to play a role in nodulation [9], although nothing is known about how such secreted proteins act. There is indirect evidence for the uptake of Nod factors into plant cells [10•,11] raising the possibility
that Nod factor recognition could occur within cells as well as at the membrane surface. Analysis of infection by rhizo-bial mutants that do not make fully substituted Nod factors has suggested that there may be different types of Nod factor receptors, one controlling early responses and another controlling invasion events [12,13]. Root chitinas-es, some of which are specifically induced during the symbiosis, can degrade Nod factors and may play an impor-tant role in the regulation of their activity [14].
Plant responses to nodulation factors
J Allan Downie
*
and Simon A Walker
†
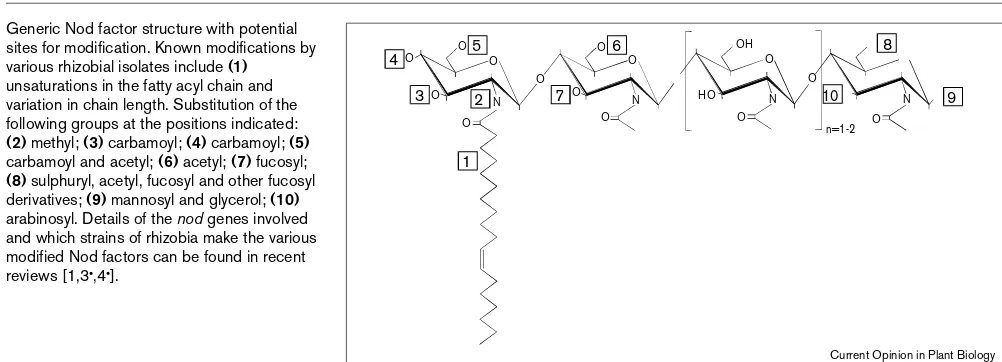
Figure 1
Generic Nod factor structure with potential sites for modification. Known modifications by various rhizobial isolates include (1)
unsaturations in the fatty acyl chain and variation in chain length. Substitution of the following groups at the positions indicated:
(2) methyl; (3) carbamoyl; (4) carbamoyl; (5)
carbamoyl and acetyl; (6) acetyl; (7) fucosyl;
(8) sulphuryl, acetyl, fucosyl and other fucosyl derivatives; (9) mannosyl and glycerol; (10)
arabinosyl. Details of the nodgenes involved and which strains of rhizobia make the various modified Nod factors can be found in recent reviews [1,3•,4•].
4 5
3 2 7
6 8
9 10
1
O
O O O
O
O O
O O
OH
HO O
O
O n=1-2 O
N N N
O N
Our review primarily addresses recent work on Nod-factor-induced changes to ion movements in and around legume root hairs and evaluates recent biochemical and genetic experiments that may lead to an understanding of the sig-nalling events several of which are summarised in Figure 2. (Note that several reports on effects of Nod fac-tors on cell division in non-legumes have recently been withdrawn [15•].) We have not described work on the
iden-tification of the many genes induced by Nod factors or on the role of plant hormones in nodule development [16,17]; recent reviews [4•,5••,18,19•,20•] describe in detail various
aspects of the plant responses to Nod factors that are not included here.
Roles for calcium
Nod factors induce a rapid and transient depolarisation of the plasma membrane of root hairs [21–23] and an associ-ated transient extracellular and sustained intracellular alkalinisation [24,25••]. A few seconds after Nod factor
exposure, there is a transient influx of Ca2+ , which is
rapid-ly followed by a Cl– efflux, and then efflux an of K+ to
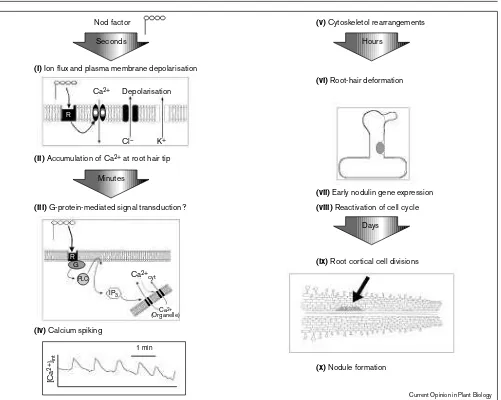
balance the charge (Figure 2i). This K+/Cl–movement and Figure 2
(i) Ion flux and plasma membrane depolarisation
(iii) G-protein-mediated signal transduction?
(iv) Calcium spiking
(v) Cytoskeletol rearrangements
(vi) Root-hair deformation
(vii) Early nodulin gene expression
(viii) Reactivation of cell cycle
(ix) Root cortical cell divisions
(x) Nodule formation
(ii) Accumulation of Ca2+ at root hair tip
Minutes
Seconds Hours
Days Nod factor
Ca2+
Ca2+ cyt
[Ca
2+
]int
Ca2+ Cl– K+
Depolarisation
(Organelle) G
IP3 PLC
R R
1 min
Current Opinion in Plant Biology
Temporal order of plant responses to Nod factors for a ‘typical’ legume. This figure draws upon data from several different legume species and the responses for any individual legume may therefore vary. No cause and effect is necessarily implied in the order of events shown. The first responses seen are (i)ion fluxes and associated depolarisation at the plasma membrane [25••], and (ii)calcium accumulation at the root hair
tip [30]. Indirect evidence suggests that (iii)a G-protein-mediated signal transduction pathway involving inositol phosphate (IP3) may be
operating [35••]; however, it is not known if this is related in any way to
the initial ion fluxes. (iv)Calcium spiking is induced [26], though it is
not known if this is a consequence of a putative G-protein pathway and/or is associated with the ion fluxes. Calcium may play a role in the
(v)subsequent changes seen in the actin cytoskeleton [32•]. The actin
cytoskeleton does appear to influence (vi)root-hair morphogenesis [33•]. (vii)Nodulation-specific gene induction is proposed to occur via
a G-protein pathway [35••] but whether calcium spiking and/or ion
fluxes play a role is not known. (viii)Reactivating the cell cycle initiates
the associated movement of H+ ions seem to account for
the membrane depolarisation.
Calcium-sensitive fluorescent dyes have been used to monitor intracellular Ca2+in root hairs. Dyes linked to high
molecular weight dextran must be microinjected but are often preferred as they remain within the cytoplasm. Acetoxymethyl esters of dyes are permeant and so can be loaded into cells by diffusion but imaging can be problem-atic because they are present at only small intracellular concentrations, diffuse out of cells and can become localised inside organelles (particularly in plant cells). Microinjection of Calcium Green/dextran into alfalfa root hairs revealed that Nod factors induced regular periodic increases in intracellular calcium that were localised most-ly around the nucleus [26]. These increases occurred about once per minute and the rapid rise and defined cut-off to the decrease in Ca2+ defined these as Ca2+ spikes
(Figure 2iv). The spiking initiated ~10 minutes after Nod factor addition and continued for at least three hours. Since Ca2+ spiking did not occur in a non-nodulating alfalfa
mutant and was not induced by Nod factors lacking a sul-phate group that is essential for alfalfa nodulation, it is very likely to form part of the normal plant response. Moreover, similar Ca2+-spiking responses have been observed in
dif-ferent spp. of Vicia, Lotusand Medicago(S Long, personal communication) and in pea root hairs (SA Walker, unpub-lished data). Ca2+ spiking plays an important role in
developmental gene regulation in mammalian cells [27•].
Analogous systems have yet to be defined in plants, although Ca2+oscillations are proposed to have a role in
regulating the opening of the stomatal guard cell aperture and are also associated with pollen tube growth [28,29].
Other studies using Ca2+-responsive dyes suggest that the
dye type and/or its method of delivery may influence the observable effects of Nod factors. Microinjection of dextran-linked dyes revealed Ca2+ spiking, whereas buffered
loading of acetoxymethyl derivatives in cowpea revealed a rapid plateau-like increase in Ca2+ at the root hair tip, but
Ca2+spiking was not observed [30]. In vetch, Nod factors
induced swelling of root hair tips and inhibited root hair growth; after ~1-1½ hrs exposure to Nod factor, root hair growth resumed and enhanced concentrations of Ca2+were
seen (as detected by the Ca2+-sensitive dye Indo 1) in the
swollen root hair tip and in the newly formed growing root hair tip [31•]. The relationships between, first,
Nod-factor-induced Ca2+ influx measured using microelectrodes,
second, the onset of Ca2+spiking and third, the
accumula-tion of Ca2+at the growing tip of the root hair have yet to be
resolved. The reasons for the different observations made by different groups could relate to differences in methodol-ogy. Microinjection may disturb the ionic equilibrium and/or the cytoarchitecture such that calcium accumulation at the tip does not occur. The age and growth phase of the root hair may be important and the temporal resolution of imaging techniques may be insufficient to allow the observation of the Ca2+spiking (discussed in [20•,31•]).
Nod factors have been shown to induce disintegration of the actin cytoskeleton and inhibit root hair tip growth within 10 mins of Nod factor addition to Phaseolus vulgaris
[32•]. At the subsequent branch-like outgrowth of the root
hairs in vetch, actin filaments reappear [33•] in the region
where there is an increased level of Ca2+(Figure 2vi) [31•]
and it was concluded that fine bundles of actin filaments promote polar growth by releasing Golgi vesicles to a vesi-cle-rich region of the growing root hair tip [33•].
Cytoskeletal changes may also be associated with the Nod factor induced reactivation of the cell cycle; outer cortical cells enter the cell cycle but do not divide, whereas inner cortical cells divide (Figure 2ix), entering the cell cycle from the G0/G1phase [34]. At a later stage in the
interac-tion, a transient disorganisation of microtubules was correlated with the appearance of Nod factors within infected cells, suggesting that Nod factors may also be involved in cytoskeletal rearrangements that occur during the differentiation of infected cells [10•].
The effects of pharmacological agents on the expression of the reporter gene MtENOD12A-GUS, which is induced in response to Nod factors, provided indirect evidence for a role for Ca2+in a G-protein-mediated signal transduction
pathway [35••]. Mastoparan, a G-protein agonist induced
MtENOD12Ain a similar way to Nod factor, whereas the G-protein antagonist, pertussis toxin, inhibited Nod factor and mastoparan-induced gene expression. Inhibitors of phospholipase C, such as neomycin, and of Ca2+
influx/release such as ethylene glycol tetraacetic acid (EGTA), La3+and ruthenium red, also blocked both the
Nod factor and mastoparan-induced gene expression. Although these results are consistent with a mammalian-type G-protein signalling pathway, coupled to the activation of phosphoinositide and Ca2+as a second
mes-senger (Figure 2iii), great care has to be taken with the interpretation of data generated by such pharmacological techniques that may be having multiple effects. The com-bination of such studies with the use of plant mutants in which signalling is blocked in the early stages should, how-ever, give an insight into the nature of the pathways involved. The identity of the Nod factor receptors are of key interest.
Nod-factor-binding proteins
Radioactively labelled LCOs were used to identify Nod fac-tor binding proteins from Medicago truncatula.A protein with a relatively low affinity (Kd = 86 nM) for the Nod factor was found but is unlikely to play a role in the symbiosis [36]. A second component, NFBS2 (Nod-factor-binding site 2), from a plasmalemma fraction [37••] has high affinity
or absence of a sulphate group on the reducing sugar did not affect Nod factor binding. This sulphate group is essential for normal Nod factor activity on Medicagospp. and so the bind-ing specificity of NFBS2 did not correspond with what would be predicted (on the basis of in vivostudies) for a puta-tive Nod factor receptor. Thus, if NFBS2 does correspond to (part of?) a receptor, the perception of the sulphate group by the Nod factor receptor would be complex, and could possi-bly involve another protein that confers additional specificity.
Isolation of proteins that bind chitin oligomers led to the identification of a Nod factor binding lectin with apyrase activity [38••]. As this lectin hydrolyses the
phosphoanhy-dride bonds of nucleoside phosphates, it was called lectin nucleotide phosphohydrolase (LNP). LNP, which was isolat-ed from the legume Dolichos biflorus, has much greater affinity for Nod factors than chitin oligomers and has greatest affinity for those Nod factors from rhizobia that can nodulate
D. biflorus. Furthermore, the apyrase activity of LNP is enhanced by Nod factors. These results, together with the observations that LNP is located on root hairs and that anti-body to LNP inhibited root-hair deformation, suggest a role for LNP in the nodule symbiosis, but as with NFBS2 a direct role for LNP in signalling has not yet been established.
Although LNP is a lectin (as defined by its sugar-binding capability), its sequence shows that it does not belong to the large family of typical legume lectins [39,40]. Several years ago it was proposed that such ‘typical’ lectins might play a role in recognition by acting as a kind of surface-binding receptor, and several studies, including addition of lectins and demon-stration of their presence on root hair tips, supported a role for lectins in the early stages of infection [40]. Furthermore, transgenic clover plants expressing a pea lectin gene can be nodulated by R. leguminosarumbv.viciae —which normally nodulates pea and not clover — and the sugar-binding domain is necessary for this activity [41]. The sugar-binding sites of such lectins, however, are not consistent with them binding Nod factors. A more recent study with transgenic plants that express an introduced lectin gene has led to a reassessment of the original hypothesis and the proposal of a different model for a role of such lectins in nodulation. Lotus corniculatus expressing a soybean lectin gene induced nodule-like outgrowths, which were devoid of bacteria, in response to the soybean symbiont Bradyrhizobium japonicum[42•]. In this
case, infection did not proceed normally and it was suggested that the introduced lectin, which was located in root-hair tips, may interact with a component of the bacterial surface poly-saccharide. It was proposed that the lectin might increase the attachment and aggregation of heterologous rhizobia (B. japonicum) at the infection site. This, in turn, could result in an increased level of Nod factors at the appropriate loca-tion, thereby potentiating a weak signal and stimulating the early stages of nodule morphogenesis, even by heterologous Nod factors. This model illustrates that there could be multi-ple inputs into recognition between rhizobia and legumes and that dissecting out the signalling pathway from other recogni-tion events may not be simple.
Legume nodulation mutants
Many nodulation-defective legume mutants have been isolated but many are in species that do not lend them-selves to gene isolation. Nevertheless, these mutants have been used to gain insight into the events that occur during nodulation. Nod–mutants of several species do not form a
mycorrhizal symbiosis [43]. The pea early nodulation tran-scripts ENOD5 and ENOD12A, which are induced in response to Rhizobium, are also induced by the endomyc-orrhizal fungus Gigaspora margarita; no expression was seen in a Nod– sym8 pea mutant when inoculated with
either symbiont [44•]. Other studies using transgenic peas
expressing an ENOD12A–GUS reporter gene demonstrat-ed that the induction of the pea ENOD12A gene is also blocked in the sym19mutant [45]. Many gene transcripts are induced early in nodulation [5••,19•,20•] and
knowl-edge of which genes are switched off in various mutants may help to determine the sequence of events that are ini-tiated after Nod factor perception, and where divergence between endomycorrhizal and rhizobial symbioses occurs.
One of the key transcripts that is induced during rhizobium–legume symbiosis is ENOD40, which is expressed prior to initiation of pericycle cell division opposite the pro-toxylem poles and is also expressed in the dividing cells [46•,47]. Transgenic legumes expressing ENOD40initiate
cor-tical cell division at multiple sites indicating that this gene influences the differentiation and division of root cells [48]. Sequence comparisons led to the realisation that ENOD40
encodes a short (10–13 residue) peptide and a conserved non-coding RNA domain that may play a regulatory role [46•,47].
Each region seems to contribute to nodule development. The conservation of this gene in non-legumes [46•] suggests that
nodule signalling may have evolved from a more ancient path-way conserved in non-legumes. Nod factor was reported to induce a M. truncatula ENOD12gene in rice [49•], although
this observation has not yet been verified by other laboratories.
Recently, there has been a focussed effort on investigating the genetics of ‘model’ legumes, an experimental counter-part to Arabidopsisfor the study of symbioses. These legumes are diploid, have relatively small genome sizes, are easy to transform and have high seed yield. Such properties lend themselves to gene-tagging strategies and positional cloning. One candidate gene for positional cloning affects the ethyl-ene sensitivity of nodulation [50]; the mutant of M. truncatula
which does not express this gene has enhanced nodulation and altered position of nodule development. These effects are due to its insensitivity to ethylene which normally down-regulates nodule development [17]. A bacterial artificial chromosome library of M. truncatula genomic DNA has been created and used to identify ethylene responsive genes [51]. Other approaches to gene identification involve a root-hair-specific cDNA library from M. truncatula [52•]. In Lotus
japonicus, an essential nodulation gene Ninhas been identi-fied by gene tagging following mutagenesis using the maize transposable element Ac[53••]. The mutant is not infected
indicating that the Ningene is unlikely to encode the prima-ry Nod factor receptor. The Ningene product is predicted to be a membrane protein and four similar Arabidopsisproteins (of unknown function) were identified in database searches. Similarity was also found with a Chlamydomonasgene product (the so-called minus-dominance protein) that regulates mat-ing type durmat-ing gametogenesis. All of these proteins share a putative DNA-binding dimerisation domain and so a regula-tory role for the Lotus nodulation gene was proposed, although the mechanism of regulation and the regulated genes are not yet known. This first characterisation of an essential nodulation gene suggests that we are now on the threshold of a phase identifying genes required to establish symbiotic interactions.
Conclusions
There is strong evidence to suggest that the early events in Nod factor recognition involve movements of ions across the plant plasma membrane, followed by the establishment of intracellular Ca2+ oscillations. Circumstantial evidence
suggests that a G-protein-type receptor that could act via an inositol-phosphate and phospholipase based signalling pathway may be involved. Much work remains to be done, however, to integrate the various observations that have been made on the legume symbiosis. The key objective in the future will be to exploit various nodulation mutants to clone and characterise genes that are essential for nodule development. The ease of the isolation (and maintenance) of legume nodulation mutants, the availability of the Nod-factor-signalling molecules, and the ability to analyse ion movements in epidermal cells mean that nodule develop-ment is a particularly good system for the analysis of a developmental pathway. Understanding how nodules are formed may well give us an insight into how other morpho-genetic events are activated in plants.
Acknowledgements
We thank NJ Brewin for his constructive comments on the manuscript and various colleagues for sending copies of recent and ‘in press’ publications. The authors are supported by the Biotechnology and Biological Sciences Research Council.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
••of outstanding interest
1. Dénarié J, Debellé F, Promé JC: Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis.Annu Rev Biochem1996, 65:503-535.
2. Kamst E, Spaink HP, Kafetzopoulos D: Biosynthesis and secretion of rhizobial lipochitin-oligosaccharide signal molecules.Subcell Biochem1998, 29:29-71.
3. Downie JA: Functions of rhizobial nodulation genes.In The
• Rhizobiaceae, edn 1. Edited by Spaink HP, Kondorosi A, Hooykaas PJJ. Dordrecht: Kluwer academic publishers; 1998:387-402.
The most comprehensive list of rhizobial Nod gene products currently avail-able. This chapter is most useful to specialists in the area.
4. Bladergroen MR, Spaink HP: Genes and signal molecules involved
• in the rhizobia-Leguminoseae symbiosis.Curr Opin Plant Biol
1998, 1:353-359.
A good overview of hormone signalling and the possible effects of flavanoids on auxins during infection.
5. Schultze M, Kondorosi A: Regulation of symbiotic root nodule
•• development.Annu Rev Gen1998, 32:33-57.
An excellent and comprehensive review of signalling between plants and bacteria focusing on plant responses.
6. Economou A, Davies AE, Johnston AWB, Downie JA: The Rhizobium
leguminosarumbiovar viciae nodOgene can enable a nodE
mutant of Rhizobium leguminosarumbiovar trifoliito nodulate vetch.Microbiology1994, 140:2341-2347.
7. Vlassak KM, Luyten E, Verreth C, van Rhijn P, Bisseling T,
Vanderleyden J: The Rhizobiumsp. BR816 nodOgene can function as a determinant for nodulation of Leucaena leucocephala,
Phaseolus vulgarisand Trifolium repens by a diversity of
Rhizobiumspp.Mol Plant–Microbe Interact1998, 11:383-392.
8. Sutton JM, Lea EJA, Downie JA: The nodulation-signaling protein NodO from Rhizobium leguminosarumbiovar viciaeforms ion channels in membranes.Proc Natl Acad Sci USA1994,
91:9990-9994.
9. Viprey V, DelGreco A, Golinowski W, Broughton WJ, Perret X:
Symbiotic implications of type III protein secretion machinery in
Rhizobium.Mol Microbiol1998, 28:1381-1389.
10. Timmers ACJ, Auriac MC, de Billy F, Truchet G: Nod factor
• internalization and microtubular cytoskeleton changes occur concomitantly during nodule differentiation in alfalfa.Development
1998, 125:339-349.
The authors of this work describe the immunolocalisation of Nod factors and tubulin, revealing a transient disorganisation of the cytoskeleton in association with Nod factor internalisation. This implies a direct associa-tion between Nod-factor-regulated symbiotic differentiaassocia-tion and the co-ordination of the cytoskeleton.
11. Philip-Hollingsworth S, Dazzo FB, Hollingsworth RI: Structural requirements of Rhizobium chitolipooligosaccharides for uptake and bioactivity in legume roots as revealed by synthetic analogs and fluorescent probes.J Lipid Res1997, 38:1229-1241. 12. Ardourel M, Demont N, Debellé FD, Maillet F, de Billy F, Promé JC,
Dénarié J, Truchet G: Rhizobium melilotilipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair-cells and induction of plant symbiotic developmental responses.Plant Cell1994, 6:1357-1374. 13. Geurts R, Heidstra R, Hadri AE, Downie JA, Franssen H, van
Kammen A, Bisseling T: Sym2of pea is involved in a nodulation factor-perception mechanism that controls the infection process in the epidermis.Plant Physiol1997, 115:351-359.
14. Schultze M, Staehelin C, Brunner F, Genetet I, Legrand M, Fritig B, Kondorosi E, Kondorosi A: Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide Nod signals.Plant J1998,
16:571-580.
15. Schell J, Bisseling T, Dülz M, Franssen H, Fritze K, John M, Kleinow T,
• Lessnick A, Miklashevichs E, Pawlowski K et al.: Re-evaluation of phytohormone-independent division of tobacco protoplast-derived cells.Plant J1999, 17:461-466.
Several previous reports suggested that Nod factors induce cell division in tobacco protoplasts. This paper retracts these observations and cites all of the work containing unreproducible data.
16. Hirsch AM, Fang Y, Asad S, Kapulnik Y: The role of phytohormones in plant-microbe symbioses.Plant Soil1997, 194:171-184. 17. Spaink HP: Ethylene as a regulator of Rhizobiuminfection.Trends
Plant Sci1997, 2:203-204.
18. Hirsch AM, Larue TA: Is the legume nodule a modified root or stem or an Organ sui generis? Crit Rev Plant Sci1997, 16:361-392. 19. Hadri AE, Bisseling T: Responses of the plant to Nod factors.In The
• Rhizobiaceae, edn 1. Edited by Spaink HP, Kondorosi A, Hooykaas PJJ. Dordrecht: Kluwer academic publishers; 1998:403-416.
An in-depth review of various aspects of plant responses to Nod factors describing the many genes induced.
20. Irving HR, Boukli NM, Kelly MN, Broughton WJ: Nod factors in
• symbiotic development of root hairs.In Cell and Molecular Biology of Root Hairs, edn 1. Edited by Ridge RW, Emons AM. Berlin: Springer-Verlag; 1999: in press.
A comprehensive review of root hair responses to Nod factors with an inter-esting discussion on potential signalling models.
22. Felle HH, Kondorosi E, Kondorosi A, Schultze M: Nod signal-induced plasma membrane potential changes in alfalfa root hairs are differentially sensitive to structural modifications of the lipochitooligosaccharide.Plant J1995, 7:939-947.
23. Kurkdjian AC: Role of the differentiation of root epidermal cells in Nod factor (from Rhizobium meliloti)-induced root-hair
depolarization of Medicago sativa.Plant Physiol1995, 107:783-790. 24. Felle HH, Kondorosi E, Kondorosi A, Schultze M: Rapid alkalinization
in alfalfa root hairs in response to rhizobial
lipochitooligosaccharide signals.Plant J1996, 10:295-301. 25. Felle HH, Kondorosi E, Kondorosi A, Schultze M: The role of ion
•• fluxes in Nod factor signalling in Medicago sativa.Plant J1998,
13:455-463.
The very earliest plant responses to Nod factors are investigated. Nod fac-tors induce a rapid influx of Ca2+that is followed by Cl–and K+efflux. These
latter ion movements can explain the Nod-factor induced depolarisation of the root hair plasma membrane.
26. Ehrhardt DW, Wais R, Long SR: Calcium spiking in plant root hairs responding to Rhizobium nodulation signals.Cell1996, 85:673-681. 27. Meldolesi J: Calcium signalling — oscillation, activation,
• expression.Nature1998, 392:863-865.
It has long been suspected that oscillations in intracellular calcium may play a role in regulating gene expression in mammalian cells. This commentary highlights the importance of two papers published in the same issue describing the first hard evidence to support this hypothesis.
28. Staxén I, Pical C, Montgomery LT, Gray JE, Hetherington AM, McAinsh MR: Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C.Proc Natl Acad Sci USA1999, 96:1779-1784. 29. Franklin-Tong VE, Drobak BK, Allan AC, Watkins PAC, Trewavas AJ:
Growth of pollen tubes of Papaver rhoeasis regulated by a slow-moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell1996, 8:1305-1321.
30. Gehring CA, Irving HR, Kabbara AA, Parish RW, Boukli NM, Broughton WJ: Rapid, plateau-like increases in intracellular free calcium are associated with nod-factor-induced root-hair deformation. Mol Plant-Microbe Interact1997, 10:791-802. 31. de Ruijter NCA, Rook MB, Bisseling T, Emons AMC: Lipochito
• oligosaccharides re-initiate root hair tip growth in Vicia sativawith high calcium and spectrin-like antigen at the tip.Plant J1998,
13:341-350.
Cytological changes induced in root hairs by Nod factors were examined by microscopy. Root hair cells with terminating growth reacquired tip-growing characteristics including a tip gradient of Ca2+and accumulation of a
spec-trin-like antigen. The results indicate that the root hairs respond to Nod fac-tors by recovering cytoplasmic polarity and exocytosis.
32. Cárdenas L, Vidali L, Domínguez J, Pérez H, Sánchez F, Hepler PK,
• Quinto C: Rearrangement of actin microfilaments in plant root hairs responding to Rhizobium etlinodulation signals.Plant Physiol1998, 116:871-877.
In this paper, researchers describe fragmentation of the Phaseolus vulgaris
root hair actin cytoskeleton upon exposure to Rhizobium etliNod factors. This was observed 5–10 minutes after Nod factor application which could correlate the response to observed changes in intracellular calcium. This work indicates that Nod factors alter the organisation of actin microfilaments in root hairs.
33. Miller DD, de Ruijter NCA, Bisseling T, Emons AMC: The role of actin
• in root hair morphogenesis: studies with
lipochito-oligosaccharide as a growth stimulator and cytochalasin as an actin perturbing drug.Plant J1999, 17:141-154.
The authors propose a model for root hair morphogenesis whereby actin fila-ments co-ordinate growth by releasing Golgi vesicles to a vesicle-rich region of the growing root hair tip. The model relies on the influence of Nod factors on root hair growth and the interference of cytochalasin D with actin function. 34. Yang WC, Deblank C, Meskiene I, Hirt H, Bakker J, van Kammen A,
Franssen H, Bisseling T: RhizobiumNod factors reactivate the cell-cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell1994,
6:1415-1426.
35. Pingret JL, Journet EP, Barker DG: RhizobiumNod factor signaling:
•• evidence for a G protein-mediated transduction mechanism.Plant Cell1998, 10:659-671.
Using a GUSreporter system to reflect early nodulin gene induction this work describes the use of various agonists and antagonists to provide indi-rect evidence for a G-protein-mediated signalling pathway operating down-stream of the Nod factor receptor.
36. Bono JJ, Riond J, Nicolaou KC, Bockovich NJ, Estevez VA, Cullimore JV, Ranjeva R: Characterization of a binding-site for chemically synthesized lipo- oligosaccharidic NodRm factors in particulate fractions prepared from roots.Plant J1995, 7:253-260. 37. Gressent F, Drouillard S, Mantegazza N, Samain E, Geremia RA,
•• Canut H, Niebel A, Driguez H, Ranjeva R, Cullimore J, Bono JJ:
Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic Nod factors in Medicagocell suspension cultures.Proc Natl Acad Sci USA1999, 96:4704-4709.
This work demonstrates that a high affinity binding protein copurifies with the plasma-membrane fraction from Medicagocell suspension cultures. Binding of LCOs with different substitutions is analysed and the presence of a sul-phate group, that is essential for nodulation, has no effect on binding affini-ty. Therefore there is some question as to whether the partially purified protein is a Nod factor membrane receptor or some other protein that hap-pens to bind LCOs.
38. Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB: A
•• Nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA1999, 96:5856-5861.
Isolation of a chitin-binding protein fortuitously led to the identification of this novel type of lectin that has both a high affinity for Nod factors and an apyrase activity. It is structurally distinct from the large family of legume lectins previously described [39,40] and a potential signalling receptor. 39. Brewin NJ, Kardailsky IV: Legume lectins and nodulation by
Rhizobium. Trends Plant Sci1997, 2:92-98.
40. Kijne JW, Bauchrowitz MA, Diaz CL: Root lectins and Rhizobia.Plant Physiol1997, 115:869-873.
41. van Eijsden RR, Díaz CL, de Pater BS, Kijne JW: Sugar-binding activity of pea (Pisum sativum) lectin is essential for heterologous infection of transgenic white dover hairy roots by Rhizobium
leguminosarumbiovar viciae.Plant Mol Biol1995, 29:431-439.
42. van Rhijn P, Goldberg RB, Hirsch AM: Lotus corniculatusnodulation
• specificity is changed by the presence of a soybean lectin gene. Plant Cell1998, 10:1233-1249.
Transfer of a soybean lectin gene enables B. japonicumto induce uninfect-ed nodules on Lotus corniculatus. A model is proposed suggesting that lectins enhance bacterial attachment to root hairs thereby enhancing Nod factor signalling by increasing localised concentration.
43. Albrecht C, Geurts R, Bisseling T: Legume nodulation and mycorrhizae formation; two extremes in host specificity meet. EMBO J1999, 18:281-288.
44. Albrecht C, Geurts R, Lapeyrie F, Bisseling T: Endomycorrhizae and
• rhizobial Nod factors both require SYM8 to induce the expression of the early nodulin genes PsENOD5and PsENOD12A.Plant J
1998, 15:605-614.
The authors find that the same plant early nodulation genes are induced by rhizobia and mycorrhizal fungi. Gene induction by both endosymbionts is blocked in a nodulation mutant of pea.
45. Schneider A, Walker SA, Poyser S, Sagan M, Ellis THN, Downie JA:
Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L).Mol Gen Genet1999, 262:1-11.
46. Kouchi H, Takane K, So RB, Ladha JK, Reddy PM: Rice ENOD40:
• isolation and expression analysis in rice and transgenic soybean root nodules.Plant J1999, 18:121-129.
The authors of this paper describe the cloning of a rice gene homologous to the early nodulin ENOD40. A promoter–GUSfusion study indicated the rice gene may play a similar role in vascular differentiation.
47. Fang YW, Hirsch AM: Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa.Plant Physiol1998, 116:53-68.
48. Charon C, Johansson C, Kondorosi E, Kondorosi A, Crespi M:
enod40induces dedifferentiation and division of root cortical cells
in legumes.Proc Natl Acad Sci USA1997, 94:8901-8906. 49. Reddy PM, Ladha JK, Ramos MC, Maillet F, Hernandez RJ, Torrizo LB,
• Oliva NP, Datta SK, Datta K: Rhizobial lipochitooligosaccharide nodulation factors activate expression of the legume early nodulin gene ENOD12in rice.Plant J1998, 14:693-702.
One of the very few papers describing activation of a legume nodulation gene by Nod factors in a transgenic non-legume.
50. Penmetsa RV, Cook DR: A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont.Science1997,
275:527-530.
51. Nam YW, Penmetsa RV, Endre G, Uribe P, Kim D, Cook DR:
Medicago truncatula and identification of clones containing ethylene-response genes.Theor Appl Genet1999, 98:638-646. 52. Covitz PA, Smith LS, Long SR: Expressed sequence tags from a
• root-hair-enriched Medicago truncatulacDNA library.Plant Physiol
1998, 117:1325-1332.
An expressed sequence tag (EST) library of root hair genes has been estab-lished and is growing rapidly. This paper describes many genes that are expressed in root hairs.
53. Schauser L, Roussis A, Stiller J, Stougaard J:A plant regulator
•• controlling development of symbiotic root nodules.Nature1999, in press.
The first paper to describe cloning of a gene essential for some of the earli-est stages in the rhizobia-legume symbiosis. Transposon-tagged mutants of