Gene expression in plants can be suppressed in a sequence-specific manner by infection with virus vectors carrying fragments of host genes. Recent developments have revealed that the mechanism of this gene silencing is based on an RNA-mediated defence against viruses. It has also emerged that a related mechanism is involved in the post-transcriptional silencing that accounts for between line variation in transgene expression and cosuppresion of transgenes and endogenous genes. The technology of virus-induced gene silencing is being refined and adapted as a high throughput procedure for functional genomics in plants.
Addresses
The Sainsbury Laboratory, John Innes Centre, Norwich, NR4 7UH, UK; e-mail: [email protected]
Current Opinion in Plant Biology1999, 2:109–113 http://biomednet.com/elecref/1369526600200109 © Elsevier Science Ltd ISSN 1369-5266
Abbreviations
PDS phytoene desaturase
PTGS post-transcriptional gene silencing
PVX potato virus X
RMD RNA-mediated defense
TBRV tomato blackring virus
TMV tobacco mosaic virus
VIGS virus-induced gene silencing
Introduction
A limiting step in genetics and molecular biology is the ability to relate genes to phenotypes and vice versa. In the shadow of genome projects this limitation is more conspic-uous than ever. Here, I describe a novel approach to bridging this gap between genetics and molecular biology. This approach is based on recent discoveries concerning plant viruses and gene silencing and is still under develop-ment. The early results, however, provide encouragement that virus-based technology will be a useful complement to existing functional genomics tools.
The most straightforward iteration of this approach is referred to as virus-induced gene silencing (VIGS) and employs virus vectors carrying elements from the exons of plant host genes. Analyses in Nicotiana species have shown that the symptoms on these plants are phenocopies of muta-tions in the corresponding host genes. For example, with tobacco mosaic virus (TMV) [1] and potato virus X (PVX) vectors [2•] carrying a fragment of phytoene desaturase (PDS) mRNA, the upper leaves of the infected plant devel-op a spectacular bleached effect (Figure 1). The cause of these symptoms is a decline in the level of the endogenous PDS mRNA leading to low levels of phytoene desaturase, a block in carotenoid production and, consequently, the absence of protection against photobleaching. In other
exam-ples VIGS has been targeted against a chelatase gene required for chlorophyll synthesis [3•] and against various transgenes [2•,3•,4••,5].
The obvious attraction of VIGS for analysis of gene func-tion is speed and ease of adaptafunc-tion to high throughput systems. If the partial sequence of a given gene is known, a virus vector construct can be assembled within two days. This construct may be infectious directly as in vitro transcripts of cDNA clones [6] or as DNA, if the virus has a DNA genome. Alternatively the cDNA of an RNA virus can be fused to a promoter that is active in plant cells. The cDNA of these constructs is infectious as directly inoculated DNA [7] or by employing an A. tumefaciens Ti plasmid vector to transfer the DNA into plant cells [8].
Fast forward genetics based on virus-induced gene silencing
David C Baulcombe
Figure 1
Nicotiana benthamiana plant exhibiting virus-induced gene silencing of phytoene desaturase. The plant was inoculated three weeks earlier with a PVX vector construct carrying a 400 nucleotide fragment of phytoene desaturase cDNA. The bleached symptoms contrast with the normal mild mosaic symptoms associated with PVX infection on this species.
After inoculation of the vector there is a delay of only 7–21 days before VIGS-related symptoms develop in the upper leaves of the infected plant. The relationship from gene to phenotype, therefore, can be established within a few weeks. In genomics jargon this strategy is referred to as reverse genetics. VIGS can also be used in a forward genetics strate-gy although the resources required are substantial — it would be necessary to construct cDNA libraries in virus vec-tor constructs and to systematically analyse large numbers of infected plants.
In the later sections of this article I assess the potential, the limitations and the practicalities of VIGS both for forward and for reverse genetics. Because the develop-ment of VIGS technology is influenced by knowledge, however, I first review the current views about the underlying mechanism.
The mechanism of VIGS
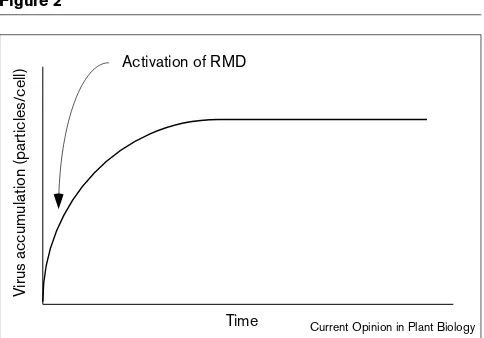
It is likely that VIGS is caused by an RNA-mediated defense (RMD) mechanism in plants against viruses [9,10]. According to this idea there is an as yet uncharac-terised surveillance system in plants that can specifically recognise viral nucleic acids and give sequence specificity to RMD. Normally, with wild-type isolates of plant virus-es, it is thought that this mechanism is activated as the virus begins to accumulate in the plant (Figure 2). Eventually, as a result of RMD, virus accumulation slows down and eventually stops.
In the situations when a genetically modified virus has similarity to a gene in the host plant, the RMD would tar-get both the viral RNA and the corresponding endogenous mRNA. VIGS would result from the targeting of the endogenous mRNA
There are two lines of supporting evidence for this model. First there is direct evidence that both RNA nepoviruses [11••] and DNA caulimoviruses can induce RMD [12,13••]. Initially the plants infected with these viruses exhibit symptoms and the viruses accumulate to a high level. However, the upper leaves of these plants emerging after systemic infection have recovered. They are symptom free and have only low levels of viral RNA. This recovery can be attributed to RMD because, in the nepovirus infected plants, there is RNA sequence-dependent resistance against viruses inoculated to these upper leaves [11••]. In the caulimovirus infected plants the recovery is probably due to RMD as there is post-transcriptional suppression of viral RNA in the symptom-free leaves.
The second supporting evidence for RMD is indirect and is based on analyses of plants exhibiting post-transcrip-tional gene silencing (PTGS) of transgenes. PTGS involves sequence-specific turnover of RNA and is one of the reasons for between-line variation in transgene expres-sion levels [14]. Lines in which a transgene is expressed at a low level include those in which PTGS is active and tar-geted against the transgene RNA. High level expressors are those in which PTGS is either weak or inactive.
The link with RMD is from the finding that PTGS is sup-pressed by virus-encoded proteins that are also suppressors of anti-viral defense in the host [15••–18••]. Presumably the suppressed anti-viral defense is the putative RMD and the suppressed PTGS was activated when the transgene or its RNA was detected by the RMD surveillance system. The suppressors of PTGS would be part of a counter defense sys-tem that allows viruses to accumulate after activation of RMD [19•]. With some viruses the final steady state level of virus accumulation would be determined by the effectiveness of the suppressors. In other instances the virus may use evasion instead of suppression as a strategy to counteract RMD.
Analysis of PTGS has revealed that there is a signal of gene silencing that can move systemically in plants [20••,21••,22•,23•]. This signal can be rationalised in terms of RMD as it would allow the plant to prevent spread of the virus. If the signal moves at the same time or ahead of the virus it would prime RMD in the cells at, or just beyond the infection front. Consequently the RMD would be activated more rapidly than in the initially infected cells and the spread of the virus would be delayed.
A final point about PTGS concerns DNA methylation. There is an association of DNA methylation and PTGS in several experimental systems [4••,24–27] and evidence that the methylation may be targeted to transgenes by RNA [4••,28]. These observations are difficult to reconcile with the proposed similarity of PTGS and RMD as most plant viruses have RNA genomes. Perhaps the DNA methylation data are an indicator that the underlying mechanism of PTGS is involved not only in anti-viral defense. It could be, for example, that antiviral defense is Figure 2
Typical time course of virus accumulation in an infected cell. The graph illustrates schematically the kinetics of virus accumulation in an infected cell. There is an initial phase of rapid replication and virus accumulation that progresses through to the final period when accumulation has been arrested. The phases shown would take place over a period of a few days, depending on the type of virus.
Time Activation of RMD
V
irus accumulation (particles/cell)
related to the mechanisms used to protect against retro-posons and other forms of mobile DNA in plants [29].
Development of VIGS technology
This relationship of VIGS, PTGS and RMD has important implications for further development of VIGS technology. It means, for example, that VIGS would be weak if the virus vector encodes a suppressor of PTGS and that the most effective VIGS vectors will be those that use evasion rather than suppression as a strategy to overcome RMD. Vectors based on potyviruses, therefore, would be poor VIGS vectors because they produce strong suppressors of PTGS [15••,16••,18••]. Cucumber mosaic virus would be unsuitable for the same reason [16••,17••]. Conversely, PVX and tomato blackring virus (TBRV) would be suitable vectors for VIGS. Neither of these viruses is able to reverse PTGS in trans-genic plants [16••,17••] and it can be assumed that they do not produce suppressors or that their suppressors are weak.
TBRV is a seed and pollen transmitted virus as a result of its ability to infect the progenitor cells of the pollen and eggs in the meristems [30]. This ability to penetrate the meristem means that TBRV would be a particularly effective vector for VIGS. Most plant viruses, including PVX, are excluded from the meristematic zone of infected plants [31] and would not be able to silence genes required for meristem identity and early in leaf and flower development. Unfortunately, as yet, there are no full length cDNAs of TBRV or related viruses that could be developed as VIGS vectors with the ability to target these important genes.
There are several other ways in which understanding of PTGS could facilitate development of VIGS technology. For example, it may be possible to enhance effectiveness of VIGS vectors by engineering increased potential to pro-duce double stranded RNA. Double stranded RNA has been implicated as an initiator of PTGS in plant systems [32•,33••,34•] and of a similar gene silencing phenomenon in nematodes [35,36•].
It may also be possible to enhance VIGS by modification of the host plant. For example, it might be expected that VIGS would be stronger in the background of host gene mutations that enhance gene silencing (egs) [37]. The products of these genes could be repressors of RMD. It might also be expected that VIGS would be stronger in plants that over-express genes required for RMD than in a wild-type host. The genes that are required for gene silencing can be recognised by analysis of the suppressed gene silencing (sgs) [38•] phenotype of mutant alleles.
Transgenic VIGS
In the applications of VIGS, as described above, the acti-vator of gene silencing is the viral RNA in the infected plant cells. The silencing effect is systemic because the virus vector is able to spread through the infected plant. In transgenic VIGS the aim is to produce an RNA that can be recognised by the RMD surveillance system. As in VIGS,
this RNA would include an element from a host sequence so that the RMD silences the corresponding gene. This RNA would not need to be infectious, however, because it would be delivered into cells by expression of a transgene.
An advantage of transgenic VIGS over ‘conventional’ PTGS using sense or antisense transgenes should be con-sistency. The transgenic VIGS constructs would be designed to produce an RNA that is detected by the RMD surveillance system and it would be expected that all, or at least most, of the lines would exhibit silencing of the tar-get gene. In PTGS it is likely that the transgene RNA is only recognised by the RMD surveillance system as a result of sequence rearrangements, DNA methylation of chromatin structures that are associated with the transgene after its introduction into the plant genome. As a result, the degree of PTGS varies greatly between lines.
Transgenic VIGS is less amenable than infectious VIGS to high throughput applications because of the transformation step. In plants that can be transformed with high efficiency, however, it is quite feasible to produce the tens of thou-sands of transformed lines that would be necessary to survey the gene function in a typical plant. Once produced, the seed from these lines would be a permanent resource that could be used in many different functional screens.
The version of transgenic VIGS corresponding most close-ly to infectious VIGS employs amplicons. Amplicons are transgenes comprising a promoter and terminator directing transcription of a modified viral vector RNA in the plant cell [39]. The modifications may include deletion or muta-tion of viral genes required for spread of the virus in the infected plant and of any other functions that are secondary to replication. Expression of the amplicon in the plant cell could activate RMD and, if there are host sequences in the amplicon construct, PTGS of the corresponding host gene.
There are several instances of amplicon transgenes although they are not always referred to as such. A PVX amplicon pro-duced consistent silencing as expected [39]. Similarly, a petunia chalcone synthase gene could be targeted by a gem-ini virus amplicon with a chalcone synthase insert [40] and a brome mosaic virus amplicon conferred virus resistance that was probably due to RMD [41]. The phenotype of ampli-cons derived from cucumber mosaic virus [42] and a severe strain of TMV, however, did not appear to involve gene silencing [43]. The differences between these amplicons may be due to the suppressors of RMD encoded in the cor-responding viral genomes — genomes producing little or no silencing may produce strong suppressors of RMD, whereas good silencers may be those that do not encode suppressors or from which the suppressor genes have been deleted.
RMD. The gene silencing from simultaneous expression of sense and antisense RNAs [33••] or from transgene con-structs with inverted repeats in the transcribed regions [32•] could both be considered in this ‘pseudoVIGS’ category .
VIGS versus other techniques of functional
genomics
Conventional approaches to forward genetics generally involve insertional mutagenesis either by T-DNA [44] or transposable elements [45,46]. These are powerful approaches that have led to many advances in gene discov-ery; however, there are several complications with insertional mutagenesis of genes required for basic cell func-tion or early development. In these instances, because the insertional mutation often results in complete inactivation of the target gene, the mutant phenotype will be embryo lethal and difficult to interpret in terms of the cellular or biochem-ical role of the gene product. Conversely, if the mutant gene is one of a family it is likely that there may be no phenotype of an insertional mutant because the functional homologues may compensate for the loss of function at the mutant locus.
VIGS and related PTGS-based approaches may circumvent these problems. For example, with multigene families, the gene silencing mechanism will target not only the precise homologue of the gene in the VIGS vector; it will also target partial homologues that vary by up to 10 or 20%. The chlorot-ic and dwarf phenotypes that result from PTGS of tobacco ribulose bisphosphate carboxylase (rubisco) is a good illustra-tion of how gene silencing can target a multigene family [47,48]. It is unlikely that a phenotype would result from insertional mutagenesis of rubisco because it is encoded in a multigene family. The precise mismatch between different tobacco rubisco genes is not known but, by comparison with other solanaceous plants, it is likely to be up to 14% [49].
When VIGS is targeted against lethal genes the phenotype is death, as in insertional mutagenesis, but the dying process may be more informative than the embryo lethal phenotype of insertional mutants. Because the VIGS is applied to mature plants and because there may be only partial suppression of the target gene, the dying process may be informative about the role of the corresponding gene products. It might be expected, for example, that death due to a block in lipid biosynthesis would be clearly different from death due to suppression of DNA synthesis or proteins required for respiration
Conclusions
VIGS vectors have been developed from the genomes of TMV, PVX and TGMV for applications in Nicotiana species. To develop these vectors for use in other plant species it will be possible to build on recent discoveries that VIGS and PTGS are related to antiviral defense in plants and that viruses produce suppressors of this antiviral defense. When these vectors are available for Arabidopsis, maize and other species it will be possible to combine inser-tional mutagenesis and VIGS to develop a powerful combined approach to functional surveys of plant genomes.
Acknowledgements
I am grateful to the Gatsby Charitable Foundation for supporting work in the Sainsbury Laboratory and to my colleagues for many stimulating discussions about the role and mechanisms of gene silencing in plants.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
••of outstanding interest
1. Kumagai MH, Donson J, Della-Cioppa G, Harvey D, Hanley K, Grill LK:
Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA.Proc Natl Acad Sci USA 1995, 92:1679-1683.
2. Ruiz MT, Voinnet O, Baulcombe DC: Initiation and maintenance of
• virus-induced gene silencing.Plant Cell1998, 10:937-946. Compares VIGS of a transgene and an endogenous gene, as in [3•]. The analysis reveals separate initiation and maintenance stages of VIGS and a surprising difference in the properties of VIGS targetted against transgenes and endogenous genes.
3. Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, • Thompson WF, Robertson D: Gene silencing from plant DNA
carried by a geminivirus. Plant J1998, 14:91-100.
Shows that a DNA geminivirus can initiate VIGS although does not rule out that viral RNA is the initiator of gene silencing. As with [2•] compares VIGS of endogenous genes and transgenes.
4. Jones AL, Thomas CL, Maule AJ: De novo methylation and
co-•• suppression induced by a cytoplasmically replicating plant RNA virus.EMBO J1998, 17:6385-6393.
Extends and establishes general importance of RNA directed methylation of DNA as reported originally in [28]. This paper used a viral system instead of a viroid as in the previous work. The paper is also important because it demon-strates VIGS in a legume. Most of the other examples are in Nicotiana species.
5. Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG:
Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance.Plant Cell 1993, 5:1749-1759.
6. Donson J, Kearney CM, Hilf ME, Dawson WO: Systemic expression of a bacterial gene by a tobacco mosaic virus-based vector.Proc Natl Acad Sci USA1991, 88:7204-7208.
7. Sablowski RWM, Baulcombe DC, Bevan MW: Expression of a flower-specific Myb protein in leaf cells using a viral vector causes ectopic activation of a target promoter. Proc Natl Acad Sci USA 1995, 92:6901-6905.
8. Turpen TH, Turpen AM, Weinzettl N, Kumagai MH, Dawson WO:
Transfection of whole plants from wounds inoculated with
Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus.J Virol Methods 1993, 42:227-240.
9. van Kammen A: Virus-induced gene silencing in infected and transgenic plants. Trends Plant Sci 1997, 2:409-411.
10. Dawson WO: Gene silencing and virus resistance: a common mechanism.Trends Plant Sci 1996, 1:107-108.
11. Ratcliff F, Harrison BD, Baulcombe DC: A similarity between viral
•• defense and gene silencing in plants.Science1997, 276 :1558-1560.
With [13••] provides an indication, in non-transgenic plants and with a wild-type virus, for an RMD that resembles PTGS.
12. Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ:
Transcriptional and post-transcriptional plant gene silencing in response to a pathogen. Science1998, 279:2113-2115.
13. Covey SN, Al-Kaff NS, Langara A, Turner DS: Plants combat
•• infection by gene silencing. Nature 1997, 385:781-782.
With [11••] provides an indication, in non-transgenic plants and with a wild-type virus, for an RMD that resembles PTGS.
14. Depicker A, Van Montagu M: Post-transcriptional gene silencing in plants.Curr Opin Cell Biol 1997, 9:372-382.
15. Kasschau KD, Carrington JC: A counter-defensive strategy of plant
•• viruses: suppression of post-transcriptional gene silencing. Cell
1998, 95:461-470.
16. Brigneti G, Voinnet O, Wan-Xiang L, Liang-Hui J, Ding S-W, •• Baulcombe DC: Viral pathogenicity determinants are suppressors
of transgene silencing in Nicotiana benthamiana. EMBO J 1998,
17:6739-6746.
This paper and the others appearing within a few weeks of each other [15••,17••,18••] provide indirect but compelling evidence that RMD is a gen-eral defense mechanism against plant viruses and that viruses have evolved counter-defense strategies.
17. Beclin C, Berthome R, Palauqui JC, Tepfer M, Vaucheret H: Infection
•• of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of non-viral (trans)genes. Virology
1998, in press.
This paper and the others appearing within a few weeks of each other [15••,16••,18••] provide indirect but compelling evidence that RMD is a gen-eral defense mechanism against plant viruses and that viruses have evolved counter-defense strategies.
18. Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Smith TH, Vance VB: A
•• viral suppressor of gene silencing in plants.Proc Natl Acad Sci USA 1998, 95:13079-13084.
This paper and the others appearing within a few weeks of each other [15••,16••,17••] provide indirect but compelling evidence that RMD is a gen-eral defense mechanism against plant viruses and that viruses have evolved counter-defense strategies.
19. Carrington JC, Whitham SA: Viral invasion and host defense:
• strategies and counter-strategies.Curr Opin Plant Biol 1998,
1:336-341.
A concise and informative review that puts RMD in the context of the vari-ous virus resistance mechanisms deployed in plants and the corresponding variety of counter-defense strategies developed by plant viruses.
20. Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H: Systemic acquired
•• silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions.EMBO J 1997, 16:4738-4745.
With [21••] the first report that gene silencing is associated with systemic signalling. The experiments involved graftings of silenced plants to non silenced plants. The systemic signal has not yet been characterised but is likely to be a nucleic acid.
21. Voinnet O, Baulcombe DC: Systemic signalling in gene silencing.
•• Nature 1997, 389:553.
With [20••] the first report that gene silencing is associated with systemic signalling. The experiments involved localised introduction of green fluores-cent protein (GFP) DNA into a GFP transgenic plant and demonstrated that local events could initiate systemic gene silencing. The systemic signal has not yet been characterised but is likely to be a nucleic acid.
22. Voinnet O, Vain P, Angell S, Baulcombe DC: Systemic spread of
• sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA.Cell 1998,
95:177-187.
Further characterises the mechanisms of systemic silencing but does not demonstrate the chemical identity of the signal.
23. Palauqui JC, Vaucheret H: Transgenes are dispensable for the RNA
• degradation step of cosuppression.Proc Natl Acad Sci USA 1998,
95:9675-9680.
Further characterises the mechanisms of systemic silencing but does not demonstrate the chemical identity of the signal.
24. English JJ, Baulcombe DC: The influence of small changes in transgene transcription on homology-dependent virus resistance and gene silencing.Plant J 1997, 12:1311-1318.
25. Sijen T, Wellink J, Hiriart J-B, van Kammen A: RNA-mediated virus resistance: role of repeated transgenes and delineation of targeted regions.Plant Cell 1996, 8:2277-2294.
26. English JJ, Mueller E, Baulcombe DC: Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes.Plant Cell 1996, 8:179-188.
27. de Carvalho F, Gheysen G, Kushnir S, Van Montagu M, Inzé D, Castresana C: Suppression of b-1,3-glucanase transgene expression in homozygous plants.EMBO J 1992, 11:2595-2602.
28. Wassenegger M, Heimes S, Riedel L, Sänger HL: RNA-directed de novo methylation of genomic sequences in plants.Cell 1994,
76:567-576.
29. Matzke MA, Matzke AJM: Epigenetic silencing of plant transgenes as a consequence of diverse cellular defence responses.Cell Mol Life Sci 1998, 54:94-103.
30. Lister RM, Murant AF: Seed transmission of nematode-borne viruses. Ann Appl Biol 1967, 59:49-62.
31. Matthews REF: Plant Virology, edn 3. San Diego: Academic Press; 1991.
32. Hamilton AJ, Brown S, Han YH, Ishizuka M, Lowe A, Solis AGA, • Grierson D: A transgene with repeated DNA causes high
frequency, post- transcriptional suppression of ACC-oxidase gene expression in tomato.Plant J 1998, 15:737-746.
Provides supporting evidence for the possible role of double stranded RNA as an initiator of gene silencing.
33. Waterhouse PM, Graham HW, Wang MB: Virus resistance and
•• gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA.Proc Natl Acad Sci USA
1998, 95:13959-13964.
Provides supporting evidence for the possible role of double stranded RNA in gene silencing. The presented data also explain why viral antisense trans-genes do not confer virus resistance. It is probably because the gene silenc-ing requires the presence of both sense and antisense RNAs.
34. Metzlaff M, O’Dell M, Cluster PD, Flavell RB: RNA-mediated RNA
• degradation and chalcone synthase A silencing in petunia.Cell
1997, 88:845-854.
Provides supporting evidence for the possible role of double stranded RNA in gene silencing.
35. Tabara H, Grishok A, Mello CC: RNA in C. elegans: soaking in the genome sequence. Science 1998, 282:430-431.
36. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC: • Potent and specific genetic interference by double-stranded RNA
in Caenorhabditis elegans.Nature1998, 391:806-811.
There is a remarkable similarity between this genetic interference in nema-todes and gene silencing in plants. See also [35].
37. Dehio C, Schell J: Identification of plant genetic loci involved in post-transcriptional mechanism for meiotically reversible transgene silencing.Proc Natl Acad Sci USA 1994, 91:5538-5542.
38. Elmayan T, Balzergue S, Beon F, Bourdon V, Daubremet J, Guenet Y, • Mourrain P, Palauqui JC, Vernhettes S, Vialle T et al.: Arabidopsis
mutants impaired in cosuppression. Plant Cell 1998, 10:1747-1757. PTGS and RMD will probably not be fully understood until mutations have been isolated and characterised in genes required for silencing. This report provides the first step to that end. It also describes plant lines that would be useful for gene over-expression.
39. Angell SM, Baulcombe DC: Consistent gene silencing in transgenic plants expressing a replicating potato virus X RNA.
EMBO J 1997, 16:3675-3684.
40. Atkinson RG, Bieleski LRF, Gleave AP, Jannsen BJ, Morris BAM: Post-transcriptional silencing of chalcone synthase in petunia using a geminivirus-based episomal vector.Plant J 1998, 15:593-604.
41. Kaido M, Mori M, Mise K, Okuno T, Furusawa I: Inhibition of brome mosaic virus (BMV) amplification in protoplasts from transgenic tobacco plants expressing replicable BMV RNAs.J Gen Virol
1995, 76:2827-2833.
42. Suzuki M, Masuta C, Takanami Y, Kuwata S: Resistance against cucumber mosaic virus in plants expressing the viral replicon.
FEBS Lett 1996, 379:26-30.
43. Yamaya J, Yoshioka M, Meshi T, Okada Y, Ohno T: Expression of tobacco mosaic virus RNA in transgenic plants.Mol Gen Genet
1988, 211:520-525.
44. AzpirozLeehan R, Feldmann KA: T-DNA insertion mutagenesis in Arabidopsis: Going back and forth.Trends Genet 1997, 13:152-156.
45. Briggs SP: Plant genomics: more than food for thought. Proc Natl Acad Sci USA 1998, 95:1986-1988.
46. Martienssen RA: Functional genomics: probing plant gene function and expression with transposons.Proc Natl Acad Sci USA 1998,
95:2021-2026.
47. Quick WP, Schurr U, Scheibe R, Schulze ED, Rodermel SR, Bogorad L, Stitt M: Decreased ribulose-1,5-bisphosphate carboxylase-oxygenase. In Transgenic Tobacco Transformed With Antisense Rbcs 1. Impact On Photosynthesis In Ambient Growth-Conditions Planta
1991, 183:542-554.
48. Hudson GS, Evans JR, Voncaemmerer S, Arvidsson YBC, Andrews TJ: Reduction of ribulose-1,5-bisphosphate carboxylase oxygenase content by antisense RNA reduces photosynthesis. In
Transgenic Tobacco Plants Plant Physiol 1992, 98:294-302.