Atherosclerosis 150 (2000) 295 – 298
A polymorphism upstream from the human paraoxonase (PON1)
gene and its association with PON1 expression
Tadashi Suehiro
a,* , Toshihiro Nakamura
a, Mari Inoue
a, Tomoko Shiinoki
a,
Yukio Ikeda
a, Yoshitaka Kumon
a, Masanori Shindo
b, Hideo Tanaka
b,
Kozo Hashimoto
aaThe Second Department of Internal Medicine,Kochi Medical School,Kohasu,Okoh-cho,Nankoku,Kochi,781-8505,Japan bCentral Pharmaceutical Research Institute,Japan Tobacco,Inc.,1-1,Murasaki-cho,Takatsuki,Osaka,569-1125,Japan
Received 25 January 1999; received in revised form 21 July 1999; accepted 3 September 1999
Abstract
Human serum paraoxonase (PON1) is an esterase that has been shown to decrease the susceptibility of lipoproteins to lipid peroxidation. We found a polymorphism of cytosine/thymidine (−108C/T, the number is from the ATG codon) in the upstream region of the PON1 gene. The luciferase activity was lower in the −108T allele than in the −108C allele. The serum PON1 concentrations in 132 normal subjects were as follows: −108CC\−108CT and\−108TT genotypes. The polymorphism upstream from the PON1 gene is associated with transcription of the PON1 gene and the serum PON1 concentration. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Paraoxonase; Lipid peroxidation; Genetic polymorphism; Coronary heart disease
www.elsevier.com/locate/atherosclerosis
1. Introduction
Human serum paraoxonase (PON1) is associated with high-density lipoprotein (HDL) [1] and has been shown to decrease the susceptibility of low-density lipo-protein (LDL) to lipid peroxidation in vitro,[2,3] which may prevent the development of atherosclerosis [4]. Recently, PON1-knockout mice were developed and were found to be more susceptible to atherosclerosis than their wild-type littermates [5]. This protein in humans has two polymorphic sites: Leu/Met at position 55 of the amino acid sequence (L/M), and Gln/Arg at position 192 (Q/R) [6]. Q/R polymorphism is associated with PON activity which is estimated using paraoxon as a substrate, and the PON activity in subjects with the QQ genotype is lower than that in subjects with the RR genotype [7]. On the other hand, L/M polymorphism is related to the serum PON1 concentrations [8]. Little information is available concerning other factors which
influence the PON1 concentration. We detected poly-morphisms in the upstream of the PON1 gene. We examined the relationship between the polymorphisms and the promoter activity of PON1 or the serum PON1 concentrations.
2. Materials and methods
Samples from 161 normal healthy subjects used in a previous study of PON1 [9], were employed in this study. The subjects visited a medical center in Kochi and were confirmed as being of normal status by phys-ical and laboratory examinations including normal rest-ing electrocardiography. No subject had diabetes mellitus, as confirmed by an oral glucose tolerance test, or a history of cardiovascular disease. As the serum from 29 subjects was completely used in previous study, only 132 of 161 samples (63 males and 69 females) were available to undergo complete detection of polymor-phisms upstream from the PON1 gene and protein concentration examination. Their mean age was 5798 (S.D.) years. The data of the 55L/M and 192Q/R polymorphisms were used from the previous study [9]. * Corresponding author. Tel.: +81-888-802343; fax: +
81-888-802344.
E-mail address:[email protected] (T. Suehiro)
T.Suehiro et al./Atherosclerosis150 (2000) 295 – 298 296
The 5% end of exon 1 of the PON1 gene was not mapped. Therefore, we applied the nucleotide number from the ATG start codon to a nucleotide position in the upstream region. PON1 in the human sera was assessed by ELISA (WAK-CHEMIE MEDICAL GMBH, Bad Soden, Germany). Briefly, ELISA plates (Corning, Corning, NY), were coated with antigen at room temperature and left overnight. The plates were blocked with 1% BSA for 1 h at 37°C. The mixtures of test sera and anti-paraoxonase antibody were incubated in a pretreated ELISA plate for 3 h at 37°C. The bound anti-paraoxonase antibody was detected with alkaline phosphatase conjugated antibody (Sigma, St Louis, MO).
We detected the 5% upstream region of the PON1 gene using a method of ‘walking upstream’ (Genome-Walker Kits, Clontech, Palo Alto, CA). We used an antisense primer whose sequence was obtained from Genebank (U55877). Variations upstream from the PON1 (−269/ +65) gene were detected by a cycle sequencing method. The DNA fragment was amplified by a PCR method using a sense primer (5% -TGGAC-TAGGCACCTATTCTC-3%) and an antisense primer (5%-GACTGGTGGTTCCTGAAGAG-3%) with genomic DNA as a template. A PCR fragment separated by electrophoresis in agarose gel was recovered and purified by an available commercial kit (GENECLEAN II Kit, BIO 101, Vista, CA). The sequence of the PCR fragment was detected using a commercial kit and analyzer (BigDye Terminator Cycle Sequencing FS Ready Reaction Kit and ABI PRISM™310 Genetic Analyzer, PE Applied Biosystems, Foster City, CA), and the same sense or antisense primer of the PCR was used for the sequencing primers.
For the firefly luciferase assay, a DNA fragment of the promoter region of the PON1 gene (−587/ −6) was amplified by the PCR method using a sense primer (5%-GGGGTACCAGCTGCATGAGGAAATG-3%) and an antisense primer (5% -TCCCCCGGGATAGA-CAAAGGGATCG-3%). The underlined area indicates the sequences added as restriction sites of Kpn1 and
SmaI, respectively. The template used genomic DNA from the −108CC or −108TT genotype whose −126 and −160 bases were guanine. The DNA fragment was introduced into the expression vector pGL3-Basic (Promega, Madison, WI), and subcloned into JM109. The introduced fragments were confirmed by the cycle sequencing method using the primers described for pGL3-Basic.
We confirmed the expression of endogenous PON mRNA in the HepG2 cell line by the RT-PCR method. The total RNA were isolated from cultured HepG2 cells and a piece of the liver obtained at an autopsy using an available kit (TRIZOL Reagent, Gaithersburg, MD). The first-strand cDNA was generated by an available kit (Ready-To-Go T-Primed First-Strand Kit,
Amersham Pharmacia Biotech, Uppsala, Sweden) and it was amplified by the PCR method (RT-PCR) using a sense primer (5%-TCAGGAACCACCAGTCTTCT-3%) and an antisense primer (5% -GGTCCCACAGCAA-CAATATC-3%), which were chosen from exon 1 and exon 6. The PCR product (475 bp) was detected in 2% agarose gel and the sequence of the product was confi-rmed by the cycle sequencing method.
Transient transfection into HepG2 cells was per-formed by a cationic lipid method using Tfx™-20 (Promega, Madison, WI). The cells were incubated in a 35 mm dish, and 13.2 ml of Tfx™-20 and 2.0mg DNA were added. After incubation for 1 h, the cells were incubated for an additional 48 h in medium with serum. The luciferase assay was performed using a commercial kit (Luciferase Assay System, Promega, Madison, WI) according to the procedure. The luciferase activities were normalized using protein concentrations (luci-ferase light units/mg protein) and expressed as relative values compared with the activity of −108C DNA. The protein concentration was measured using the Protein Assay Kit (Bio-Rad Lab, Richmond, CA).
All data were presented as the mean9S.D. The distribution of two PON1 genotypes of – 108C/T and 192Q/R were estimated by contingency table analysis. Comparison of variables among groups was performed using one-way ANOVA and each comparison was esti-mated by Fisher’s protected least significant difference. Comparison of two groups was estimated by the un-paired t-test.
3. Results
Three polymorphisms were detected in the upstream fragment (−269/ +65) as follows:−108C/T, −126G/ C, and −160G/A. The −126G/C and −160G/A polymorphisms were almost linked. All −160GG and −160AA homozygotes showed −126GG and −126CC homozygotes, respectively. Only 1.8% of −160GC heterozygotes showed −126GG homo-zygotes. The frequencies of the −126C allele and −160A allele were low (0.09 and 0.10, respectively). The frequencies of −108CC, −108CT, and −108TT genotypes were 25.8, 44.7, and 29.5%, respec-tively. Allele frequencies of C and T were 0.48 and 0.52, respectively. Table 1 shows the distributions of −108C/T and 192Q/R. There was a significant rela-tionship between the −108C/T and 192Q/R polymor-phisms. The frequency of the 192RR genotype in the −108TT genotype was higher than that in the −108CC genotype.
T.Suehiro et al./Atherosclerosis150 (2000) 295 – 298 297
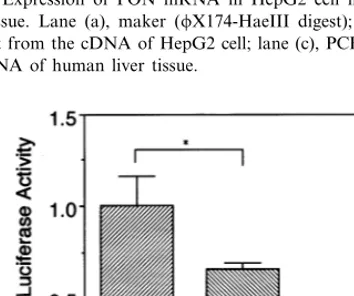
four times, and the mean activity of the −108T allele was 0.67 – 0.77-fold lower than that of the −108C allele.
The mean concentration of serum PON1 was as follows: −108CC\−108CT\−108TT genotype
Fig. 3. The serum concentrations of PON1 in each genotype of the
−108G/C polymorphism of the PON1 gene. ANOVA is PB0.01.
*PB0.05, **PB0.001, Fisher’s test. Table 1
Distributions of−108C/T and 192Q/R polymorphisms of the PON1 gene in normal healthy subjectsa
PON−108C/T,n(column %)
CC CT TT Total
PON192R/Q,n(column %)
6 (10.2) 1 (2.6)
QQ 7 (20.6) 14
23 (67.6) 39 (67.1)
RQ 16 (41.0) 78
40 22 (56.4)
14 (22.7) RR 4 (11.8)
132 Total 34 (100.0) 59 (100.0) 39 (100.0)
a
x2:PB0.001.
(Fig. 3). The level in the −108CC was about 1.2-fold higher than that of the −108TT genotype. The 192Q/ R, −126G/C, or−160G/A polymorphism revealed no relationship to the serum PON1 concentration. How-ever, there was a significant relationship between the 55L/M polymorphism and concentrations. The number of subjects with LL, LM or MM genotype was 115, 17, or 0. The concentration was higher in LL genotype than in LM genotype (175945 vs. 152935 mg/ml, respectively, PB0.05). Among the group of LL geno-type, the concentration was also related to the −108C/ T polymorphism. The concentration of −108CC, −180CT, or −108TT genotype in the group was 204946, 177942, or 154938 mg/ml, respectively (PB0.001, ANOVA).
4. Discussion
PON1 exhibits several enzymatic activities associated with artificial substrates [10], but its physiological func-tion is unknown except for a protective effect against lipid oxidation of LDL or HDL [2,3,11]. The enzyme activity for an artificial substrate may not always indi-cate the physiological effect of PON1. The 192Q/R polymorphism may be involved in CHD, and the RR is related to an increased risk of CHD [12,13]. However, PON activity in the RR genotype is higher than that in the QQ genotype. The serum PON1 concentration is related to 55L/M polymorphism although the 192 poly-morphism had little impact on the concentration [8]. The relationships among enzyme activities, mass con-centrations, and clinical diseases have not been established.
The −108C/T variation is a common polymorphism at least in Japanese populations, although the polymor-Fig. 1. Expression of PON mRNA in HepG2 cell line and human
liver tissue. Lane (a), maker (fX174-HaeIII digest); lane (b), PCR product from the cDNA of HepG2 cell; lane (c), PCR product from the cDNA of human liver tissue.
Fig. 2. The relative luciferase activities of the upstream region in the
−108C allele and the −108T allele of the PON1 gene. *PB0.001,
T.Suehiro et al./Atherosclerosis150 (2000) 295 – 298 298
phism has been reported in the Genebank (accession No. AF051133) from Switzerland. PON1 is dis-tributed in many tissues including the liver [10], and the HepG2 cell line expresses the PON1 mRNA [14]. Therefore, we studied the reporter gene assay of PON1 gene using HepG2 cells, and the results showed that the −108C/T polymorphism was associ-ated with the transcription of PON1. Moreover, the relationship between the genotypes and serum concen-trations of PON1 suggested that the polymorphism was also associated with the serum concentration. Al-though the distributions of 54L/M and −108C/T polymorphisms were linkage disequilibrium, the con-centration of PON was related to the −108C/T poly-morphism not only in all subjects, but also in the group of 54LL genotype. Therefore, the relationship between the serum concentration and the −108C/T polymorphism might be independent of the 54L/M polymorphism.
The sequence of the −108C allele (ggggcggggc) showed a consensus sequence of the Sp1 binding site [15]. The region of polymorphism may be involved in the transcription through Sp1. However, the promoter activity of the −108T allele by the luciferase assay decreased by 0.7-fold compared with that of the − 108C allele. Therefore, the region only partly regu-lates PON1 transcription. Our reporter gene assay was estimated without any stimulation. The promoter activity may be influenced by many factors in vivo, and changes in various conditions. In addition, the inserted DNA of the PON1 upstream region was within 600 bp. There may be other stronger regula-tory sites further upstream. The relationships among other sites in the PON1 upstream region and the −108C/T polymorphism should be examined.
Recently, Mackness et al. showed that the PON1 QQ-HDL was the most efficient at protecting LDL against oxidative modification and PON1 RR-HDL was the least efficient [16]. The 192Q/R polymor-phism may affect the ability of HDL to protect LDL from oxidation. However, Cao et al. could not confirm lower activity of the R allele towards lipid peroxides [17]. Our result showed that the −108C/T polymorphism was linked to the 192Q/R polymor-phism. The association of 192Q/R polymorphism with CHD may be partly reflected by the −108C/T poly-morphism. Furthermore, the relationship between polymorphisms of the PON1 gene and CHD must be clarified.
References
[1] Blatter M-C, James RW, Messmer S, et al. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45. Identity of K-45 with paraoxonase. Eur J Biochem 1993;211:871.
[2] Mackness MI, Arrol S, Durrington PN. Paraoxonase prevents accumulation of lipoperoxides in low-density lipoprotein. FEBS Lett 1991;286:152.
[3] Mackness MI, Arrol S, Abbott C, et al. Protection of low-den-sity lipoprotein against oxidative modification by high-denlow-den-sity lipoprotein associated paraoxonase. Atherosclerosis 1993;104: 129.
[4] Mackness MI, Arrol S, Abbott CA, et al. Is paraoxonase related to atherosclerosis? Chem Biol Interact 1993;87:161.
[5] Shih DM, Gu L, Xia Y-R, et al. Mice lacking serum paraox-onase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998;394:284.
[6] Adkins S, Gan KN, Mody M, et al. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet 1993;52:598.
[7] Humbert R, Adler DA, Disteche CM, et al. The molecular basis of the human serum paraoxonase activity polymorphism. Nature Genet 1993;3:73.
[8] Blatter Garin M-C, James RW, Dussoix P, et al. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J Clin Invest 1997;99:62.
[9] Ikeda Y, Suehiro T, Inoue M, et al. Serum paraoxonase activity and its relationship to diabetic complications in patients with non-insulin-dependent diabetes mellitus. Metabolism 1998;47: 598.
[10] La Du BN. Human serum paraoxonase/arylesterase. In: Kalow W, editor. Pharmacogenetics of Drug Metabolism. New York: Pergamon, 1992:51.
[11] Aviram M, Rosenblat M, Bisgaier CL, et al. Paraoxonase in-hibits high-density lipoprotein oxidation and preserves its func-tions. A possible peroxidative role for paraoxonase. J Clin Invest 1998;101:1581.
[12] Ruiz J, Blanche´ H, James RW, et al. Gln-Arg192 polymorphism of paraoxonase and coronary heart disease in type 2 diabetes. Lancet 1995;346:869.
[13] Serrato M, Marian AJ. A variant of human paraoxonase/ arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest 1995;96:3005.
[14] Navab M, Hama-Levy S, Van Lenten BJ, et al. Mildly oxidized LDL induces an increased apolipoprotein J/paraoxonase ratio. J Clin Invest 1997;99:2005.
[15] Briggs MR, Kadonaga JT, Bell SP, Tjian R. Purification and biochemical characterization of the promoter-specific transcrip-tion factor, Sp1. Science 1986;234:47.
[16] Mackness B, Mackness MI, Arrol S, et al. Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipo-protein oxidative modification. FEBS Lett 1998;423:57. [17] Cao H, Girard-Globa A, Berthezene F, et al. Paraoxonase
protection of LDL against peroxidation is independent of its esterase activity towards paraoxon and is unaffected by the QR genetic polymorphism. J Lipid Res 1999;40:133.
.