Development of multiplex-PCR assay for rapid detection of
Candida spp
.
Ni Made A. Tarini,I Mardiastuti H. Wahid,2 Fera Ibrahim,2 Andi Yasmon,2 Samsuridjal Djauzi3 1 Department of Clinical Microbiology, Medical Faculty, University of Udayana, Bali, Indonesia
2 Department of Microbiology, Faculty of Medicine, University of Indonesia, Jakarta, Indonesia
3 Allergic-Immunology Division, Department of Internal Medicine, Faculty of Medicine, University of Indonesia – Ciptomangunkusumo Hospital, Jakarta, Indonesia
Abstrak
Tujuan Uji biokimia untuk identifi kasi Candida spp. memakan waktu dan menunjukkan hasil yang tidak dapat ditentukan. Metoda deteksi spesifi k untuk antibodi, antigen dan metabolit Candida spp. memiliki sensitivitas dan spesifi sitas yang rendah. Pada penelitian ini kami mengembangkan metoda diagnostik cepat, Uji Reaksi Rantai Polimerasa Multipleks (Multiplex-PCR) assay untuk identifi kasi Candida spp.
Metode Lima isolate Candida spp. dibiak, diidentifi kasi menggunakan uji germ tub, dan kit API® 20 C AUX (BioMerieux®
SA). Selanjutnya, DNA dipurifi kasi dengan kit QIAamp DNA mini (Qiagen®) untuk uji Multiplex-PCR.
Hasil Batas deteksi DNA dengan uji Multiplex-PCR assay dari C. albicans, C. tropicalis, C. parapsilosis, C. krusei dan C. glabrata berturut-turut adalah 4 pg, 0,98 pg, 0,98 pg, 0,5 pg and 16 pg Uji ini lebih sensitif daripada biakan karena Multiplex-PCR dapat mendeteksi 2.6-2.9 x 100 CFU/ml, sementara biakan hanya 2.6-2.9 x 102 CFU/ml. Kesimpulan Multiplex PCR menunjukkan sensitivitas yang lebih tinggi dari biakan. Uji ini dapat direkomendasikan sebagai uji yang sensitive dan spesifi k untuk identifi kasi Candida spp. (Med J Indones 2010; 19: 83-7)
Abstract
Aim Candida spp. infection commonly occur in immunocompromised patients. Biochemical assay for identifi cation of Candida spp. is time-consuming and shows many undetermined results. Specifi c detection for antibody, antigen and metabolites of Candida spp. had low sensitivity and specifi city. In this study, we developed a rapid diagnostic method, Multiplex-PCR, to identify Candida spp.
Methods Five Candida spp. isolates were cultured, identifi ed with germ tube and API® 20 C AUX (BioMerieux® SA) kit.
Furthermore, DNA was purifi ed by QIAamp DNA mini (Qiagen®) kit for Multiplex-PCR assay.
Results DNA detection limit by Multiplex-PCR assays for C. albicans, C. tropicalis, C. parapsilosis, C. krusei and C.
glabrata were 4 pg, 0,98 pg, 0,98 pg, 0,5 pg and 16 pg respectively. This assay was also more sensitive than culture in that Multiplex-PCR could detect 2.6-2.9 x 100 CFU/ml, whereas culture 2.6-2.9 x 102 CFU/ml.
Conclusion Multiplex-PCR is much more sensitive than culture and thus, can be recommended as a sensitive and
specifi c assay for identifi cation of Candida spp.(Med J Indones 2010; 19:83-7)
Key words:Candida spp., multiplex-PCR
Correspondence email to: [email protected]
Recently, many fungal infections were caused by a group of Candida spp. The fungal infection constitutes an opportunistic infection that commonly causes candidiasis in immunocompromise patients. Fungal infections may be superficially acute or chronic infections, relating to mouth, oropharyngeal and esophagus mucosal skin, and systemic infection.1-3
Identifi cations of Candida spp. by conventional culture technique followed by biochemical tests are time consuming. In addition, biochemical tests sometime fail to identify the microorganisms, while API 20C AUX assay takes long time to complete, around 2-3 days. Dealing with this assay, Gundes et al. reported that 12% of tested isolates need additional treatment and
0.8% are not well identifi ed.4 The assays for detection of specifi c antibody, antigen and metabolite of Candida have already been developed, however their sensitivity and specifi city are very low. The reason why an antibody detection is not applicable for candidosis detection, is that colonization of Candida spp. in intestinal tract and other sites of the body induce antibody response of either immunocompetent or immunocompromise people.5
an early alternative therapy can be administered.6,7 Polymerase Chain Reaction (PCR) is a nucleic acid-amplifi cation technique. This technique is very sensitive and can amplify a small amount of DNA target, even for a single cell DNA.8,9 Multiplex-PCR is a rapid, sensitive and specifi c method which combining many specifi c-species primers in one PCR tube. Therefore, it can be used to detect the causative agents simultaneously.10,11 In this study we developed Multiplex-PCR, a rapid diagnostic assay for identifi cation of Candida spp.
METHODS
Isolates of
A. Candida spp
Candida albicans ATCC 10231 and Candida glabrata isolates were obtained from Clinical Microbiology Laboratory, Faculty of Medicine, University of Indonesia (FMUI). Candida tropicalis, Candida parapsilosis, and Candida krusei were obtained from Parasitology Laboratory, FMUI. All isolates were identifi ed by using the germ tube technique and API® 20 C AUX (BioMerieux® SA) according to the manufacturer’s instruction.
DNA extraction B.
DNA genome of Candida spp was extracted and purifi ed by QIAamp DNA mini (Qiagen®) Kit. Brie
fl y, single colony of fungi was taken by stick applicator sterile and inoculated into 5 ml YPD medium. The medium was incubated on orbital incubator at 30oC and 250 rpm for 16-24 h. Spectrophotometer was used to measure the cell density. Three milliliter medium containing cell density of 1.2 x 108 cells/ml (OD
600 = 5–10) was centrifuged at 7.500 rpm for 5 min. Pellet was suspended with 600
μl Sorbitol Buffer and 200 U lyticase (Sigma®). The mixture was then incubated at 30oC for 30 min, and centrifuged at 7.500 rpm for 5 min. For obtaining DNA genome, pellet was extracted by QIAamp DNA mini (Qiagen®) following manufacturer’s instruction. Fifty micro liters of fi nal elute containing DNA was stored at– 20oC until used. 12
Multiplex-PCR C.
Multiplex-PCR was performed with a total volume of 30 μl, consists of: 1x hotstar buffer solution (Qiagen®), 1 mM MgCl2 solution (Qiagen®), 0,2 mM dNTP Mix (Biogen), 0,4 μM each of ITS1 and ITS2 primers, 1.2 μM each of CA3 and CA4 primers, 0,025 U of
HotStar taq DNA polymerase (Qiagen®), and 2 μl (2-6 ng/reaction) of DNA template. The primer sequences used in this reaction were universal primers (ITS1 TCCGTAGGTGAACCTGCGG-3’] and ITS2 [5’-GCTGCGTTCTTCA TCGATGC-3’], and specifi c primers (CA3 [5’-GGTTTGCTTGAAAGACGGTAG-3’]) and CA4 [5’–AGTTTGAAGATATACG TGGTAG-3’]).(10) Products were amplifi ed using the following conditions (PCR thermal cycler MJ Mini PCR System BioRad): 950C for 15 min, then 40 cycles of 940C for 30 sec, 600C for 60 sec, and 720C for 45 sec followed by one cycle of 720C for 5 min. For annealing temperature, primer was reacted with all fi ve DNA templates (C. albicans, C. tropicalis, C. parapsilosis, C. krusei and C. glabrata) at 60.0, 60.6, 61.5, 62.8, 64.4, 65.6, 66.6 and 67.0oC. This reaction was aimed to know the optimal annealing temperature of all fi ve DNA templates. For detection of amplifi cation results, the PCR products were run on 9% polyacrylamide gel at 100 volt for 1.5 h. Gel was stained with 0.04% ethidium bromide for 5 minutes. The DNA bands were detected by Gel Doc XR with ultra violet transiluminator. A positive test result was claimed if a DNA band corresponding to each Candida species was detected on polyacrylamide gel, except for C. albicans in that two DNA bands had to be detected.
Sensitivity D.
Minimal DNA detection of Multiplex-PCR was tested towards all fi ve DNA templates of Candida spp. A series of the two-fold DNA dilution (0.5 - 0.00025 ng/μl) in 1x PBS was conducted. Sensitivity of Multiplex-PCR assay was performed by comparing it with conventional culture. Ten-folded dilutions (stock: 2 x 105 cells) of C. albicans in 1x PBS were carried out. For each dilution, 200 μl suspension was applied for culture in SDA medium and another 200 μl was tested by Multiplex-PCR. After cells isolated from the medium, it were incubated at 35oC for 48 h, the number of fungal colonies were then calculated (CFU/ml).
Specifi city E.
RESULTS
1. Annealing Temperature of Multiplex-PCR Assay
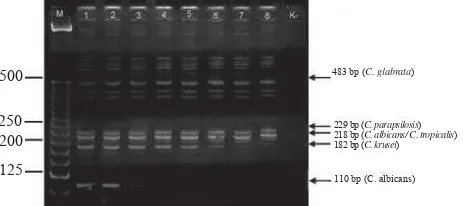
The optimal temperature was determined by visually examination of the DNA band intensity on polyacriyamide gel. The optimal annealing temperature was defi ned as the annealing temperature that yielded the highest specifi c PCR products corresponding to particular species of Candida spp. At annealing temperature of 61.5oC, a 110 bp-DNA band intensity corresponding to C. albicans showed a faded DNA band, and it disappeared at annealing temperature of 62.8oC (Figure 1). For C. tropicalis, C. parapsilosis, C. krusei and C. Glabrata, their specifi c DNA bands showed the considerable DNA intensities at annealing temperature of 60.0 and/or 60,6oC. Therefore, we decided that the optimal annealing temperature of primers used in this assay for all fi ve DNA was 60.0oC (Figure 1)
2. Sensitivity
In this study, DNA detection limits by Multiplex-PCR assay for C. albicans, C. tropicalis, C. parapsilosis, C. krusei and C. glabrata were 4.0, 0.98, 0.98, 0.5, and 16 picogram (pg) respectively (data not shown). To know the interferences among primers, we compared the results of PCR reactions using all primers (mixture of universal primer and C. albicans specifi c primers) and only a universal primer pair. These studies showed that DNA detection limit using a universal primer pair was 0.4 pg, while mixture of all primers was 4 pg (data not shown). It explained that the mixture of primers in a Multiplex-PCR system decreased by ten-fold DNA detection.
Moreover, sensitivity of Multiplex-PCR system was also analyzed by comparing it with conventional culture
(Table 2). The Multiplex-PCR assay was able to detect C. albicans cells at dilution of 2 x 102 corresponding to 2.6 – 2.9 x 101 CFU/ml, while culture could only detect
C. albicans cells at dilution of 2 x 103 corresponding to 2.6 – 2.9 x 102 CFU/ml.These results showed that the Multiplex-PCR assay developed in this study was ten times more sensitive than conventional culture for identifi cation of Candida spp.
3. Specifi city
Multiplex-PCR assay developed in this study generated negative results against the following fungus: A. niger, A. fl avus, and A. fumigatus. Besides fungus, this assay also yielded negative results against a selection of bacteria, including S. pneumoniae, S. alpha haemolyticus, S. beta haemolyticus, S. gamma haemolyticus, S. aureus, S. epidermidis, Morraxella, Diptheroid, H. infl uenzae, and N. sica.
DISCUSSION
Primers employed for determination of Multiplex-PCR were similar to primers used by Chang, et al. namely, universal fungi-primers (ITS1 and ITS2) and specifi c primers for Candida albicans (CA3 and CA4). ITS1 and ITS2 primers are targeted to the conserved parts of 18S rDNA and 5.8S rDNA. These universal primers produced varies DNA fragments of Candida spp. Candida albicans specifi c primers, CA3 and CA4, amplifi ed ITS2 region of Candida albicans.7 Based on Fujita, et al, ITS region was located between 18S gene and 28S rRNA. ITS was divided into ITS1 (located between 18S and 5.8S rRNA genes) and ITS2 (located between 5.8S and 28S rRNA genes). The sequence differences in ITS1-5.8S rDNA-ITS2, ITS1 and ITS2 regions among fungus lead the regions to be used as 500
250 200
125
483 bp (C. glabrata)
229 bp (C. parapsilosis) 218 bp (C. albicans/ C. tropicalis) 182 bp (C. krusei)
110 bp (C. albicans)
Figure 1. Annealing temperature of Multiplex-PCR assay. M: DNA ladder. Line 1-8: annealing temperatures at 60.0, 60.6, 61.5, 62.8, 64.4, 65.6, 66.6 and 67.0oC re-spectively. K-: PCR negative control
Fungi Dilution in 1x PBS
Conventional culture* (duplo)
Number of colonies (CFU/ml) (duplo)
Multiplex-PCR*
I II I II
C. albicans 2 x 105 + + TD TD +
2 x 104 + + 2.6 x
103
2.9 x 103 +
2 x 103 + + - - +
2 x 102 - - - - +
2 x 101 - - - -
-2 x 100 - - - -
targets for fungal detection and identifi cation.13 Lau, et al. reported that universal primers targeted to ITS1 region for identifi cation of pathogenic fungi were an ideal approach, because there were multi copies (approximately ≥ 100 copies) of this region in single fungal genome; therefore increasing the sensitivity assay. Also, this region contained the high variable sequences that were useful for species identifi cation.14
In this research, sample was inoculated on YPD medium and then incubated for overnight (16-18 hours) before DNA extraction. Lysis of fungal cell with special lysis enzyme to produce spheroplast was an important step for extraction of fungal DNA. For obtaining a maximum DNA concentration, suspension containing fungal cells should be incubated overnight until achieving 105 fungal cells before the addition of lysis enzyme. 15
Concerning about the sensitivity, Multiplex-PCR assay was able to detect C. albicans cells at dilution of 2 x 102 corresponding to 2.6 – 2.9 x 101 CFU/ml. According to Einsele, et al., detection limit of PCR using primers with target sequence of 18S rDNA was 1 CFU/mL blood.7 In this research, primers with the same sequence target of 18S rDNA were also applied, but there were result differences as reported by Einsele, et al. These differences may be caused by technique applied. Einsele and colleagues were applied a hybridization method that was more sensitive than conventional PCR, while in this study we only applied conventional PCR.
The result of this research was also not agreed with study reported by Chang, et al. They reported that the detection limit of Multiplex-PCR using same primers as performed in this study was is 20 CFU/ml.10 This differences in specimen type may be infl uence the result obtained between Chang, et al study and ours. In this case, Chang used blood, whereas in our study we use throat swab. As reported by Fredricks, et al. cited by Pryce, et al., blood contains inhibition factors such as hemoglobin and lactoferin In addition, specimens containing BACTEC and BacT/ALERT compounds are also not applicable for PCR due to the present of inhibition factors such as sodium polyanetholesulphonate (SPS).16
Multiplex-PCR sensitivity towards DNA Candida spp were varied, sensitivity of Multiplex PCR towards DNA Candida glabrata was lower compare to others. Sensitivity of fungal molecular determination was 10 fg, however, 1 – 10 fungal cells per mL.17 According to Khlif, et al., determination of Candida spp using PCR
method can detect less than 1 genome cell per 1 ml sample, he applied Nested PCR Method and used ITS1-ITS4 primers. Amplifi cation in Nested PCR method was applied twice, the fi rst amplicon was amplifi ed further to increase sensitivity.18 The differences are in the primer used, between pair of ITS4 and ITS1-ITS2 was not affecting the sensitivity, however size or length of the amplicon detected was affected. ITS1-ITS4 primers amplifi ed regions 3’ 18S which bind to ITS1 region, 5.8S rDNA that bind to ITS2 region and 28S rDNA region.13,18 In accordance to Chang, et al., sensitivity towards DNA Candida albicans obtained was 4 pg and this result was the same with the sensitivity found in this research. Based on Jaeger, et al., detection limit of fungal cell DNA using molecular method obtained was 10 pg and predicted similar to 100 cells, in which 1 Candida albicans cell containing of 37 fg DNA.10,19 Primer sensitivity towards Candida spp DNA in their research is varied. According to Jordan, et al., the different observed can be due to some factors, including primer design applied or sensitivity different of cell wall chitin shell against fungi lysis enzyme.15
Inconclusions, multiplex-PCR (2,6-2,9 x 100 CFU/ml) was more sensitive, accurate and rapid, compared to culture (2.6-2.9 x 102 CFU/ml) in identifying Candida spp. with sensitivity towards DNA of C. albicans ATCC 10231, C. tropicalis and C. parapsilosis, C. krusei and C. glabrata were 4 pg, 0.98 pg, 0.5 pg, and 16 pg, respectively. While Multiplex-PCR DNA control positive concentration for C. albicans ATCC 10231, C. tropicalis and C. parapsilosis, C. krusei and C. glabrata obtained were 0.6 ng/μl, 0.2 ng/μl, 0.4 ng/
μl, and 1.4 ng/μl, respectively. In the future, this assay would be evaluated using suffi cient clinical specimens to study the validity of this assay
REFERENCES
Kwon-Chung KJ, Bennet JE. Candidiasis. In: Medical 1.
Mycology. Philadelphia-London: Lea & Febiger. 1992, p 280-326.
Richardson MD, Warnock DW. Superfi cial Candidosis. In: 2.
Fungal Infection Diagnosis and Management. 3rd Edition.
Blackwell Publishing. 2003, p 109-27.
Syarifudin PK. Epidemiologi Kandidosis. J Mikol Ked 3.
Indon. 2002;3:28-30.
Wins W, Allen S, Janda W, Koneman E, Procop G, 4.
Schreckenberger P, Woods G. The Laboratory Identifi cation of Yeast. In: Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 6th Edition. Lippincott Williams
Xiang H, Xiong L, Liu X, Tu Z. Rapid Simultaneous 5.
Detection and Identifi cation of Six Species Candida Using Polymerase Chain Reaction and Reverse Line Hybridization assay. J Microbiol Methods. 2007;69:282-7.
Posteraro B, Sanguinetti M, Masucci L, Romano L, Morace 6.
G, Fadda G. Reverse Cross Blot Hybridization Assay for Rapid Detection of PCR-Amplifi ed DNA from Candida
Species, Cryptococcus neoformans, and Saccharomyces cerevisiae in Clinical Samples. J Clin Microbiol. 2000;38:1609-14.
Einsele H, Hebart H, Roller G, Loffl er J, Rothenhofer I, 7.
Muller CA, et al. Detection and Identifi cation of Fungal Pathogens in Blood by using Molecular Probes. J Clin Microbiol 1997;35:1353-60.
Strachan T, Read AP. Human Molecular Genetic. 3
8. rd Edition.
New York: Garland Publishing. 2004.
Richarson MD, Warnock DW. Laboratory Diagnosis of 9.
Fungal Infection. In: Fungal Infection Diagnosis and Management. 3rd Edition. Blackwell Publishing; 2003. p.
14-28.
Chang HC, Leaw SN, Huang AH, Wu TL, Chang TC. 10.
Rapid Identifi cation of yeasts in Positive Blood Cultures by a Multiplex PCR Method. J Clin Microbiol. 2001;39(10): 3466-71.
Luo G, Mitchell TG. Rapid Identifi cation of Pathogenic 11.
Fungi Directly from Cultures by Using Multiplex PCR. J Clin Microbiol. 2002;40:2860-65
QIAamp
12. ® DNA Mini Kit Handbook. QIAGEN. Australia/
USA.2003.
Fujita SI, Senda Y, Nakaguchi S, Hashimoto T. Multiplex 13.
PCR Using Internal Transcribed Spacer 1 and 2 Regions for
Rapid Detection and Identifi cation of Yeast Strains. J Clin Microbiol. 2001;39:3617-22.
Lau A, Chen S, Sorrel T, Carter D, Malik R, Martin P, et 14.
al. Develpment and Clinical Application of a Panfungal PCR Assay to Detect and Identify Fungal DNA in Tissue Specimens. J Clin Microbiol. 2007;45(2):380-5.
Jordan JA, Durso MB. Rapid Speciation of the Five Most 15.
Medically Relevant Candida species Using PCR Amplifi cation and a Micro titer Plate-Based Detection System. Molecular Diagnostic. 1996;1:51-7.
Pryce TM, Palladino S, Price DM, Gardam DJ, Campbell 16.
PB, Christiansen KJ, et al. Rapid Identifi cation of Fungal Pathogens in BacT/ALERT, BACTEC, and BBL MGIT Media Using Polymerase Chain Reaction and DNA Sequencing of the Internal Transcribed Spacer Regions. Diagnostic Microbiology and Infectious Disease. 2006;54: 289-97.
Hopfer RL, Amjadi D. Molecular Diagnosis of Fungal 17.
Infections. In: Calderone RA, Cihlar RL, editors. In: Fungal Pathogenesis Principles and Clinical Applications. New York: Marcel Dekker, Inc. 2002, p. 649-65.
Khlif M, Sellami H, Sellami A, Makni F, Cheikhrouhou F, 18.
Chelly H, et al. Detection and Identifi cation of Candida sp.
by PCR in Candidemia Diagnosis. Journal de Mycologie Medicale. 2007;17:256-60.
Jaeger EE, Carrol M, Choundhury S, Dunlop AS, Towler 19.