COMPREHENSIVE STUDY ON THE CHROMOSOME OF
HYLOBATIDAE GENUS Hylobates
AND Symphalangus
HERY WIJAYANTO
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
DECLARATION AND REFERENCES CITED
I hereby declare that the dissertation entitled “Comprehensive Study on the Chromosome of Hylobatidae Genus Hylobates and Symphalangus” is my own research as part of the Hylobatids Projects, Research Cooperation between the Primate Research Institute, Kyoto University, Japan and the Primate Research Center, Bogor Agricultural University, Indonesia, and has never been published, nor submitted to other universities before. All of the references cited from other sources were written inside the text and listed at the references in the last part of this dissertation.
Bogor, Agustus 7, 2005
COMPREHENSIVE STUDY ON THE CHROMOSOME OF HYLOBATIDAE GENUS Hylobates AND Symphalangus
Hery Wijayanto1,2,4*), Sri Supraptini Mansjoer1,2), Hirohisa Hirai3), Dondin Sajuthi1,2)
1. Primatology Study Program, Postgraduate School Bogor Agricultural University, Bogor, Indonesia, Jl. Lodaya II/5 Bogor 15161, Indonesia.
2. Primate Research Center, Bogor Agricultural University, Bogor, Indonesia, Jl. Lodaya II/5 Bogor 15161, Indonesia.
3. Primate Research Institute, Kyoto University, Inuyama, Aichi 484-8506, Japan. 4. Veterinary Anatomy, Gadjah Mada University, Jl. Olahraga, Bulaksumur,
Yogyakarta, 55281 Indonesia
Abstract
The study was done to investigate the chromosomes treats of Hylobates (agile gibbon) and Symphalangus (siamang). Blood samples were collected from Sumatra (agile gibbons and siamangs) and Kalimantan (agile gibbons). The blood samples then were cultured for chromosome staining. The results showed that the chromosomes of agile gibbons were too sensitive for temperature treatment, and intensive attention must be given to this aspect during handling. Using C-band after DAPI (4’-6-diamidino-2-phenylindole) staining techniques, it was found that agile gibbons have a specific chromosome banding pattern, with i (interstitial), t (terminalis), and c (pericentromeric) heterochromatic bands in both (short and long) of the chromosome arms. More intense observations using Fluorescence In-situ Hybridization (FISH) technique, it was also discovered that a Whole Arm Translocation between chromosome 8 and 9 (WAT 8/9) phenomenon occurred in Sumatran, but never in Kalimantan agile gibbons, and this proved to be very useful as a tool to differentiate between Sumatran and Kalimantan species and will give a significant contribution to avoid hybridization caused by missed species identification. Above all, the application of genetic aspects in cases such as chromosome identification in the ex-situ breeding colonies and rehabilitation centers is strongly suggested as the last barrier before releasing captive or confiscated animals into the wild. This program will not only avoid physically reducing animal populations in the natural habitat, but more importantly, to guarantee their genetic purity in their own wild habitat.
Title : COMPREHENSIVE STUDY ON THE CHROMOSOME OF HYLOBATIDAE GENUS Hylobates AND Symphalangus
Author : Hery Wijayanto
NRP : B 066010021
Study Program : Primatology
Approved by advisors:
Dr.Ir.Hj. Sri Supraptini Mansjoer
Prof. Dondin Sajuthi, DVM, MST. Ph.D. Prof. Hirohisa Hirai, Ph.D.
Acknowledged by,
2. Chairperson of Primatology Study Program
3. Dean of Graduate School
Dr.Ir.Hj. Sri Supraptini Mansjoer Prof.Dr.Ir. Syafrida Manuwoto, M.Sc.
COMPREHENSIVE STUDY ON THE CHROMOSOME OF
HYLOBATIDAE GENUS Hylobates
AND Symphalangus
HERY WIJAYANTO
This Dissertation is one of requirements needed to obtain a Doctorate degree
at the Primatology Study Program
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
CURRICULUM VITAE
The author was born in Palembang, South Sumatera on June 28, 1963 from Mr. Supadi and Mrs. Sujanti. He graduated from Senior High School (SMAN I Sragen) in 1982, and in the same year enrolled at the Faculty of Veterinary Medicine of Gadjah Mada University, and became one of the education staff in the same institution since 1990 up to now. His Master degree was obtained from the Graduate Veterinary Science Program, Gadjah Mada University in 1996. He became vice director of the Veterinary Medicine Non-degree Program in the same institution from 1995-2001.
In 2001, the author was accepted as a Doctorate Candidate in the Primatology Study Program, Graduate School, Bogor Agricultural University. He became involved in the Hylobatids Projects (a joint research between the Primate Research Institute, Kyoto University and the Primate Research Center, Bogor Agricultural University, Indonesia) from 2002 up to now.
Science experiences
Visiting researcher: Yamaguchi University, Japan (1999); Gifu University, Japan (2000); Primate Research Institute, Kyoto University (2003, 2004, 2005).
Presenter at the Annual Meeting of the Veterinary Sciences, Osaka Prefecture University, Japan (1999).
Courses:
PREFACE
“Comprehensive Study on the Chromosome of Hylobatidae Genus
Hylobates and Symphalangus”iscomposed ofthree chapters, initiated by staining technique, which was already modified to obtain a better result in C-banding of Hylobates chromosome (Article 1); the chromosome identification and traits in the Hylobates and Symphalangus (Article 2); and finished by Whole Arm Translocation of chromosome 8 and 9 in the Sumatran agile gibbon (Article 3).
The dissertation is part of the Hylobatids Project, a Joint Cooperation Research between the Primate Research Center, Bogor Agricultural University, Indonesia and the Primate Research Institute, Kyoto University, Japan, which has been implemented over the last three years. Blood samplings for genetic analysis of agile gibbon were done in several places e.g. Central Kalimantan, South Kalimantan, West Sumatera, Indonesian Safari Park (Taman Safari Indonesia), Ragunan Zoo, Japan Monkey Center, and Primate Research Institute, Kyoto University, Japan. The third Article was published in Chromosome Research 13: 123–133, 2005, while the second Article is in printing process at the same journal. I am thank full for support of the Rector of Gadjah Mada University, Yogyakarta; Rector, Dean and Staff of the Graduate School, Primatology Study Program, and Primate Research Center, Bogor Agricultural University; Primate Research Institute, Kyoto University, Japan; BPPS-DIKTI; Conservation and Natural Resources Bureaus (BKSDA) of Central Kalimantan, South Kalimantan, and West Sumatera; Indonesian Safari Park, Cisarua, Bogor; Ragunan Zoo, Jakarta; Japan Monkey Center; Inuyama, Japan; and Dean and Staff of the Faculty of Veterinary Medicine, Gadjah Mada University, Yogyakarta. My deepest gratitude are expressed to the advisory committee for their assistance in this dissertation; Dr. Ir. Sri Supraptini Mansjoer, Prof. drh. Dondin Sajuthi, MST, Ph.D., and Prof. Hirohisa Hirai, Ph.D. My appreciations go to Prof. Dr. Ir. Hadi S Alikodra and Dr. Noviar Andayani, M.Sc., for their kind suggestions in the final examination. My special appreciations go to drh. Ikin Mansjoer, M.Sc., for his kindness to review this dissertation, and Dr. Kim Hoe Su from Pusan University for his kind provided of TSPY probe. My deepest feelings of gratitude go to my lovely wife and kids, and the extended families of Supadi and Yahoedi Prijohamidjojo for their never ending supports and sacrifices.
This publication is dedicated to the conservation program of the small apes. It is a small effort in the prevention of the extinction of the small apes and proof that this genetic identification technique can be used to maintain their purity.
Bogor, August 2005
TABLE OF CONTENT
Page
LIST OF FIGURE……….
INTRODUCTION ………
Background……… Purposes of the researches..……….. References ………
MATERIAL AND METHODS………
Sampling Locations……… Nomenclature for classifying C-banded chromosomes……….. PRINS localization of telomeric DNA………. Data analyses……….
References……….
Differentiation and Confirmation of C-heterochromatin Traits Using DAPI/C-band Sequential Staining in Chromosome of the Agile Gibbon (Hylobates agilis) and the Siamang (Symphalangus syndactylus) ………
Abstract………. Comparison between DAPI- and C-bands ……….. Discussion ……… Conclusion……… Acknowledgement ……… References………
Patterns of C-heterochromatin and telomeric DNA in two
representative groups of small apes, the genera Hylobates and
Introduction………
A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features……….……
Abstract………..
Introduction……… ………
Materials and methods……….………..
Samples, chromosome preparation and DNA extraction ………. Chromosome painting analysis ……… DNA analysis ………... Results and discussions………..…………
Formation of WAT8/9 ………. Incidence of WAT8/9 ………..
Molecular genetic relationship between Sumatran and Bornean gibbons………..
Perspective and conclusion ………...
Acknowledgement ……… ………
References……….
GENERAL DISCUSSION..………..………
Chromosome karyotype as the landmark for species identification…………..
Fluorescence in situ hybridization (FISH) technique for speciesIdentification………..
DNA relationship of Sumatran and Kalimantan agile gibbon………...
LIST OF FIGURE
Page MATERIALS AND METHODS
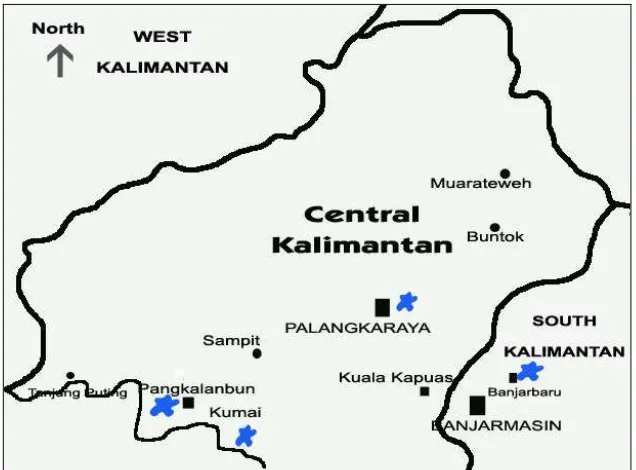
Figure 1. Map of Sumatera and Kalimantan where the samples were collected from the origin habitat………
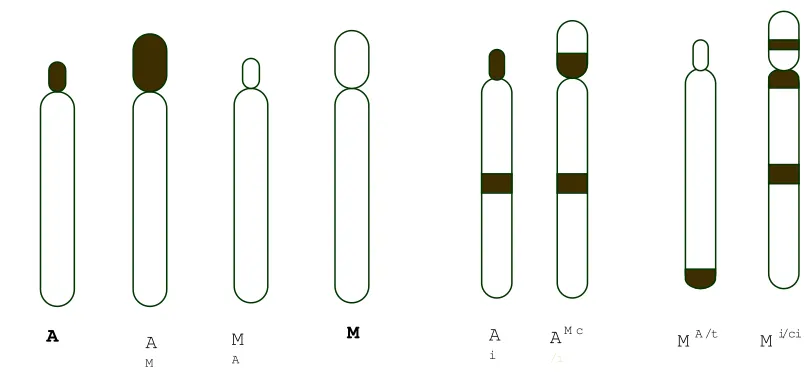
Figure 2. Nomenclature of C-banding pattern according to TAM system (Imai 1991 and Imai et al. 2001), modified by Hirai (Hirai et al. 2002)………..
Figure 3. The schematic diagram of research works……… 1. Differentiation and Confirmation of C-heterochromatin Traits Using
DAPI/C-band Sequential Staining in Chromosomes of the Agile Gibbon (Hylobates agilis) and the Siamang (Symphalangus syndactylus)
Figure 1. Chromosomes of agile gibbons. DAPI staining and C-band after DAPI-staining ……….
Figure 2. Chromosomes of siamangs. DAPI staining and C-band after DAPI staining ………
Figure 3. Ideogram explains the expression of DAPI and C-band sequential staining in chromosomes of agile gibbons (Hylobates) and siamangs (Symphalangus) ………...
Figure 4. C-banding after DAPI-staining. Standard method of BSG (Sumner 1970) and modified protocol of BSG applied to Hylobates chromosomes ………
Figure5. Schematic diagram of C-banding pattern of Hylobates chromosomes and Symphalangus ………. 2. Patterns of C-heterochromatin and telomeric DNA in
two representative groups of small apes, the genera
Hylobates and Symphalangus
Figure 1. C-banded metaphase spreads of a male agile gibbon (Hylobates agilis) and a female siamang (Symphalangus syndactylus) stained by a direct method... Figure 2. C-band karyotyping of agile gibbons. ……….
Figure 3. Comparison of DAPI-band and C-band in a metaphase spread of siamang by DAPI/C-sequential staining……… Figure 4. Localization of telomeric DNA of agile gibbons and siamang
labeled by a PRINS technique with a primer (CCCTAA)7………
3. A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features
Figure 1. Partial map of Indonesia showing original localities of gibbons of known origin on the islands of Sumatra and Borneo, and historical sea level changes of Sunda land at the last glacial period ……… Figure 2. Chromosome painting analysis of agile gibbons using human
chromosome painting probes HSACs 9, 17, and 22. ……… Figure 3. Schematic pathway of chromosome changes in agile gibbons
and expected heterozygotic patterns ………
Figure 4. Schematic relationships between Sumatran (UN/AG) and Bornean (AL) agile gibbons, and Müller’s Bornean gibbons (MU), showing the network of TSPY gene sequences, genetic distance between groups (arrows) calculated with alleles of microsatellite DNA loci, and incidence of WAT8/9 of males ………...
48 49 53
INTRODUCTION
Background
The Hylobatidae is a family of the Asian small apes, which are endangered (except Symphalangus). They consist of four genera, Bunopithecus, Hylobates, Symphalangus, and Nomascus. They are categorized as critically endangered
primate species, and are included in Appendix I (threatened species) of CITES (Convention on International Trade in Endangered Species of Wild Fauna and Flora), together with great apes and other monkeys (Soehartono and Mardiastuti 2002). Massive deforestation is the main cause of their threatened status, and has destroyed much of their habitat and reduced their population. Conservation programs are urgently needed to prevent their extinction. However, species or subspecies identification of the small ape have become a serious problem at zoological institutions due to the morphological similarities which they possess, making determination difficult, even for an expert. To solve this problem, a new technique with species-specific identification availability is urgently needed. The technique must be simple, cheap, and applicable for small laboratories with limited facilities.
One of a few very good landmarks for genus and species identification in small apes is chromosome karyotypes, because in the higher eukaryotic, the chromosome are composed of a number of repetitive DNAs, which have played an important role in constructing the chromosome shape and may have affected the divergence of chromosomes by mutation (Zimmer et al. 1980, reviewed in Hirai et al. 1999). Small apes show very different banding patterns and numbers of
separating the four genera of small apes, that is, Bunopithecus (2n=38), Hylobates (2n=44), Symphalangus (2n=50), and Nomascus (2n=52) (reviewed in Geissmann 1995). Based on the facts above, and to provide the identification tool needed for conservation using chromosome land marking, the present serial researches were conducted.
Two of four genera of Hylobatidae exist in Indonesia, Hylobates and Symphalangus. The Hylobates group consists of six species, H. agilis, H. lar, H.
muelleri, H. klossii, H. molloch, and H. pileatus, while Symphalangus has only one
species, S. syndactylus, and is the only gibbon species that occurs sympatrically with other gibbon species (H. agilis and H. lar), throughout its whole distribution area (Geissmann 1995). Hylobates agilis, the agile gibbon, is found in Thailand, Indonesia (Sumatra and Kalimantan), and Malaysia, while S. syndactylus (siamang) is found in Indonesia (Sumatra) and Malaysia. They live in the upper canopy of the forest, feeding on fruits, leaves and insects. Hylobates spend most of their lives in the trees, and rarely descend to the ground (Marshall & Sugardjito 1986). Lesser apes in the family Hylobatidae are generally small. The average weight of Hylobates agilis is about 4-5.8 kg (females) and 7 kg (males). Agile gibbons have a
variety of different colors, starting from black, brown, to light brown/yellowish and reddish-brown. Males have white eyebrows and cheeks, while females only possess white eyebrows. Hylobates agilis, like other gibbons, have extremely long arms and fingers and have no tail. This forelimb adaptation supports in brachiation, a locomotion that consists of hanging from branches and swinging from tree to tree (Marshall& Sugardjito 1886).
the pair' s territory. Hylobates agilis defend their territory by "singing". In the early morning, great calls can be heard throughout the upper canopy. These singing bouts are often duets and are a way of claiming home territory. When singing is not enough to keep intruders away, both male and female gibbons will chase the intruder away (Napier 1972; Brockelman & Gittins 1984; Marshall & Sugardjito 1986; Geissmann 1995).
So far, their taxonomy is limited understood, caused by the inconsistencies of the genetic results from different researchers and methods, amid their conservation status, which are getting critique from time to time. Many conservation programs are starting in many places, but some very important biological databases for the success of the Hylobatids conservation program has not been completely uncovered yet, e.g. chromosome data. Unlike other species where their chromosomes have similar morphology among closely related species, hylobatid karyotypes differ by their extensive chromosome reshuffling, and show very rare similarities from one species to others. Various conflicting interpretations in gibbon phylogeny indicate that they may have diverged within a relatively short evolutionary time, resulting in poor correlations between genetic (DNA sequence), cytogenetic (chromosome differentiation), and morphological divergence (Müller et al. 2003; Hirai et al. 2005).
Hirai et al. (2003), using FISH probe of human chromosomes (HSACs) painting probes 22 (CP5622-red and -green, Qbiogene), 17 (CP5617-green), and 9 (CP5605-red), found that a Whole Arm Translocation (WAT) phenomenon occurred in chromosome eight and nine of the Sumatra agile gibbons, but not in those of Kalimantan origin. This finding becomes a very good tool to differentiate between Sumatra and Kalimantan agile gibbons. However, since the WAT phenomenon above resulted from captive animals with limited numbers, a continuing study was conducted (Hirai et al. 2005) to reconfirm it, by collecting the samples from known origin animals and captive animals around their natural habitat. The latter study supports the first hypothesis, and again, WAT was only found in Sumatra agile gibbons. Since to observe WAT the karyotypes identification is focusing only on chromosome 8 and 9, where the WAT occurs (Article No. 3), this study then was extended to identify the entire agile gibbon chromosome (Article No. 2), which very important for cytogenetic and evolutionary studies.
The agile gibbon chromosome data then were compared with those of siamang (the genus Symphalangus) that have distinct C-bands to infer chromosome differentiation of small apes. To completing our data, we extend our observation on the telomeric sequence both of this small apes using primed in situ labeling (PRINS) technique. It is very important to identify both of these genera precisely, because the main point of the modern conservation is how to maintain the genetic purity of the species.
Purposes of the researches
The researches were conducted to figure out the complete identification
of C-band heterochromatin and traits of Hylobates and Symphalangus
chromosome. According to the best of the investigator’s knowledge, mapping
of C-banding pattern of agile gibbons is not provided yet, because of the
complexity of C-banding pattern on agile gibbon chromosomes but, will be
very useful to determine the exact sub-species of agile gibbon. This finding will
be very important in the success of agile gibbon conservation.
The final goal is to provide a tool for genetic identification of Hylobatid
species, partly, when morphologically identification is difficult to accomplish
due to their similarities. In the future, species identification based on genetic
markers could be a method of choice for identification to avoid hybridization
in captivity, because species of the genus Hylobates are readily interbred in
captivity, though small hybrid populations now are known at points of contact
between some of these species in the wild (Hirai et al. 2003).
References
Arnold N, Stanyon R, Jauch A, O’Brien P, Weinberg J (1996) Identification of complex chromosome rearrangements in the gibbon by fluorescent in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet Cell Genet 74: 80–85.
Brockelman and Gittins (1984) reviewed in agile gibbon
http://members.tripod.com/ uakari/ hylobates_agilis.html, downloaded 9 Des 2002.
Geissmann T (1995) Gibbon systematics and species identification. IntlZoo News 42: 467–501.
property of a whole-arm translocation (WAT) between chromosomes 8 and 9 of agile gibbons (Hylobates agilis). Chromosome Res 11: 37-50. Hirai H, Taguchi T, Godwin AK (1999) Genomic differentiation of 18S ribosomal
DNA and â-satelite DNA in the hominoid and its evolutionary aspects,
Chromosome Res. 7: 531-540.
Hirai H, Wijayanto H, Tanaka H, Mootnick AR, Hayano A, Perwitasari D,
Iskandriati D & Sajuthi D (2005) A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features, Chromosome Res 13: 1–11.
Jauch A, Wienberg J, Stanyon R, Arnold N, Tofanelli S, Ishida T, Cremer T (1992) Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Natl Acad Sci USA 89: 8611-8615.
Koehler U, Bigoni F, Weinberg J, Stanyon R (1995) Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30: 287–292.
Marshall J, Sugardjito J (1986) Gibbon systematics. In: DR Swindler, J Erwin eds., Comparative Primate Biology, Vol. 1: Systematics, Evolution, and Anatomy. New York: Alan R. Liss, pp. 137–185.
Müller S, Hollatz M, Wienberg J (2003) Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113: 493-501.
Napier P (1972) Monkeys and Apes, New York, Grosset and Dunlap Publisher.
Nie W, Rens W, Wang J, Tang F (2001) Conserved chromosome segments in Hylobates hooloc revealed by human and H. leucogenys paint probes. Cytogenet Cell Genet 92: 248–253.
Soehartono T, Mardiastuti A (2002) CITES: Implementation in Indonesia. Jakarta, Nagao Natural Environmental Foundation.
MATERIAL AND METHODS
Sampling Locations
Most of the samples were obtained from captive gibbon near their
original habitat. Blood samplings were taken with the permission of the
Indonesian Research Authority/Lembaga Ilmu Pengetahuan Indonesia (LIPI)
and Indonesian Forestry Protection and Nature Conservation/Perlindungan
Hutan dan Pelestarian Alam (PHKA), with the assistance from the rangers of
the local Natural Resources Conservation Office/Balai Konservasi Sumber
Daya Alam (BKSDA) in West Sumatra, Central Kalimantan, and South
Kalimantan.
Sumatra and Kalimantan were selected as the locations of blood
sampling of gibbons since both of the islands are the natural habitat of agile
gibbons. H. agilis albibarbis (Kalimantan); and H. agilis agilis and H. agilis
ungko (Sumatra) belong to the same species H. agilis. Recently, a subspecies H.
agilis albibarbis was promoted to be separated as a species rank (Groves 2001).
Methods
Blood sampling
Blood collection was conducted as described before (Hirai et al. 1998).
Whole blood (less than 1 ml/kg) from gibbons was collected using heparinized
syringes under anesthetized conditions with ketamine hydrochloride. The
methods of anesthesia (7.5 mg ketamine/kg body weight by an intramuscularly
injection)) and blood sampling (2 ml/kg body weight) were in accordance with
the guidelines for the care and use of primates of the Primate Research
Institute, Kyoto University; KUPRI; Primate Research Institute, Kyoto
University 2002 and the Primate Research Center, Bogor Agricultural
University 2003. Blood samples were cultured at the Primate Research Center;
preparations were made after 70 h culture, as previously described (Hirai et al.
2003).
Figure 1. Map of West Sumatera and Central Kalimantan. Blue stars are places where the samples were collected in the origin habitat. West Sumatera, H. agilis agilis (upper), Central Kalimantan (bellow), H. agilis albibarbis, and South Kalimantan, H. muelleri
Chromosome preparation
Chromosome preparation was conducted at the Laboratory of Virology, Primate Research Center, Bogor Agricultural University. The technique was adopted from Hirai et al. (1998). One milliliter of whole blood was cultured in 9ml PRMI 1640 medium containing the following agents: 20% FCS, mitogen (10 µg/ml phytohemaglutinin), 50-ug/ml streptomycin, and 50 units penicillin. The culture was incubated in 5% CO2 incubator, 37oC for 70 hr. The culture was treated with 50
Laboratory techniques
C-banding and DAPI staining were performed following the standard procedure adopted in the Department of Cellular and Molecular Biology, Primate Research Institute, Kyoto University (Hirai 2001). DAPI staining was done prior to the banding staining and proved very useful to clarify the C-banding pattern of each chromosome.
DAPI-C band sequential staining
Each chromosome was first identified using DAPI (4,6-diamidino-2-phenylindole) staining prior to C-banding in order to describe C-band traits of all chromosomes. DAPI fluorochrome has a higher affinity with A+T-rich DNA than with G+C-rich DNA (reviewed by Sumner 1990), so that it can be used to define the C-bands. Brighter fluorescence is positive DAPI and contains A+T rich DNA, while dull band is negative DAPI, G+C rich DNA.
DAPI staining was done by denaturing slides in 2xSSC pH 12.5 for 4.5 min followed by 70% and 99.5% alcohol dehydration for 5 min each, and covering with anti-fade solution containing DAPI (50 ng/mL). A cover slip was attached after drying and immersing in BI buffer (0.1 mol/L sodium bicarbonate and 0.1 % IGEPAL (Sigma) for 5 min.
tap water at the intervals between each step. After drying, the stained slide was covered with a mounting reagent (Malinol) and a cover slip, and the C-banded chromosomes were analyzed for chromosome spreads examined DAPI bands. Treatment temperature 53°οC showed more stable stainability for C-banding after DAPI staining rather than 55oC used for direct C-banding, being also needed to get clearer standard C-band patterns (for the detailed method, see the article text).
Nomenclature for classifying C-banded chromosomes
To classify C-banded karyotypes of small apes, we used Imai’s TAM system (Imai 1991; Imai et al. 2001) slightly modified by Hirai et al. (2002), which is very useful for identifying chromosomes with terminal (t), interstitial (i), and pericentromeric (c) C-bands.
Figure 2. Nomenclature of C-banding pattern according to TAM system (Imai 1991 and Imai et al. 2001), modified by Hirai (Hirai et al. 2002). Imai classify chromosome into four shapes, A, AM, MA and M. To describe the agile gibbon chromosome, which showed i (interstitial), t (terminal), and c (pericentromeric) C-band, the nomenclature system then was modified using term of cit. A slash (/) between superscripts letters is for indicating which arm (short or long) has the cit (Hirai et al. 2002). That is, the left side is in the short arm and the right in the long arm
A A
M
M
A
M A
i
AM c /i
Basically, C-banded chromosomes were divided into four types distinguished with chromosome morphology and C-band system: A and AM (‘A’ chromosome) are chromosomes with the totally heterochromatic short arm and euchromatic long arm, while MA and M (‘M’ chromosome) are chromosomes with euchromatic short and long arms. Each chromosome type can be characterized in more detail in terms of ‘c’, ‘i’, and ‘t’ band. A slash (/) between superscripts letters is for indicating which arm (short or long) has the cit (Hirai et al. 2002). That is, the left side is in the short arm and the right side is in the
long arm.
PRINS localization of telomeric DNA
Telomeric sequences (T2AG3)n were located using PRINS (primed in situ) technique as described by Hirai (2001). Reaction solution was prepared following the manufacturer’s protocol (Boehringer Mannheim, Dig-PRINS reaction kit). The results were observed and saved using a Zeiss Axiophoto epifluorescence microscope with a cooled CCD camera system (Photometrics, PXL) connected to computer (G4, Apple Macintosh) running IPLab software (Scanalytics).
Natural habitat Captive animal
Blood culture DNA extraction
Chromosome analysis DNA analysis
Sequencing TSPY gene
C-band traits & pattern
Data analyses
Chromosome data were analyzed descriptively, while TSPY divergence was confirmed by a genetic distance analysis with micro satellite DNA fragments. The FST value was calculated and a 5000 time permutation test for its significance was performed by AMOVA using data from 195 alleles of 12 micro satellite loci. References
Groves CP (2001) Primate Taxonomy, Smithsonian Institution Pess, London. Hirai H (2001) Relationship of telomere sequence and constitutive heterochromatin
in the human and apes as detected by PRINS. Meth Cell Sci 23: 29-35. Hirai H, Hirai Y, Kawamoto Y, Endo H, Kimura J, and Rerkamnuaychoke W
(2002) Cytogenetic differentiation of two sympatric tree shrew taxa found in the southern part of the Isthmus of Kra. Chromosome Res 10:313-327.
Hirai H, Mootnick AR, Takenaka O, Suryobroto B, Mouri T, Kamanaka Y, Katoh A, Kimura N, Katoh A, and Maeda N (2003) Genetics mechanism and property of a whole-arm translocation (WAT) between chromosomes 8 and 9 of agile gibbons (Hylobates agilis). Chromosome Res 11: 37-50. Hirai H, Hasegawa Y, Kawamoto Y, Tokita E (1998) Tandem Duplication of
Imai HT (1991) Mutability of constitutive heterochromatin (C-bands) during eukaryotic chromosome evolution and their cytological meaning. Jpn J Genet 66: 635-661.
Imai HT, Satta Y, Takahata N (2001) Computer simulations of mammalian and hymenopteran chromosome evolution based on the minimum interaction theory. J theor Biol 210: 475-497.
Differentiation and Confirmation of C-heterochromatin Traits Using DAPI/C-band Sequential Staining in Chromosomes of the Agile Gibbon (Hylobates agilis) and the Siamang (Symphalangus syndactylus)
Hery Wijayanto1,2,4*), Sri Supraptini Mansjoer1,2), Yuriko Hirai3), Diah Perwitasari 2), Diah Iskandriati2), Hirohisa Hirai3), Dondin Sajuthi2)
1) Primatology Study Program, Postgraduate School Bogor Agricultural University, Bogor, Indonesia, Jl. Lodaya II/5 Bogor 15161, Indonesia.
2) Primate Research Center, Bogor Agricultural University, Bogor, Indonesia, Jl. Lodaya II/5 Bogor 15161, Indonesia.
3) Primate Research Institute, Kyoto University, Inuyama, Aichi 484-8506, Japan. 4) Veterinary Anatomy, Gadjah Mada University, Jl. Olahraga, Bulaksumur,
Yogyakarta, 55281 Indonesia
*)
Correspondence
Abstract
Wide varieties of staining techniques have been used in chromosome studies and produce characteristic banding patterns of chromosomes that are important clues for research of chromosome evolution. One of those techniques, the DAPI (4’-6-diamidino-2-phenylindole)/C-band sequential staining technique is method of choice for chromosome identification in species and locations of constitutive (C) heterochromatin. The DAPI fluorescence banding technique is simple and is applicable in the limited laboratory facilities as well and does not comparatively damage chromosomes, and if it is combined with C-banding technique, thereby, provides a very useful method to clarifying the heterochromatin traits. The present report uses a C-banding technique with barium hydroxide which stains in particular C-heterochromatin. Application of C-band staining in chromosomes of the agile gibbons, a species of lar-group gibbons, need a slightly modified technique compared to the standard protocol, since the animal has more sensitive stainability for C-banding than other hominoids. Our trials showed that temperature plays a critical role in obtaining good C-bands in the chromosomes of lar-group gibbons. Reducing incubation temperature (from 60 to 53oC) of 5% Ba(OH)2 for 7 minutes and 2xSSC for 25 minutes followed by soaking in 4%
Giemsa solution at room temperature for 30 minutes resulted in more contrast banding than using the standard protocol. More over, the DAPI/C-band sequential staining technique revealed that C-heterochromatin of lar-group gibbon and siamang contain G+C and A+T rich DNA segments, respectively.
Introduction
Many methods of chromosome staining have so far been established in cytogenetic studies. Chromosomes rearrangements occurring during evolution can be tracked back by chromosome staining, and allow us to determine the break points precisely. One of the useful techniques is C-banding method, detecting constitutive (C-) heterochromatin. Heterochromatin was firstly defined by Heitz (1928) as chromatin that did not decondense at the end of the telophase, but instead remained compact throughout the interphase, and was found to be condensed even at the beginning of the prophase (Sumner 2003). Many methods have been tried to demonstrate C-bands on chromosomes during several decades. For instance, C-band produced by DNase have been used on sectioned material by Yamasaki, however, inconsistencies on C-banding patterns some times occurred using such method, Katho et al. demonstrated heterochromatic bands in chromosomes of the Indian muntjac, which were different from the C-bands shown by the BSG technique, Dev et al. used incubation in formamide solution which were adopted by Marshall to
induce C-bands, unfortunately this protocol only worked in the chromosomes of mice, but not in chromosomes of rat, man, or hamster, after trypsin G-banding (Yamasaki 1961; Katho et al. 1974; Dev et al. 1972; Marshall 1975, reviewed in Sumner 1990). To solve the inconsistency of C-bands results, Sumner (1972) introduced the BSG (Barium hydroxide/Saline/Giemsa) method, which has became the standard method for producing C-bands on plant and animal chromosomes (Sumner 1990). However, standard protocol of C-banding, which was applied for staining agile gibbon chromosome in fact did not work well. In the present study, an experiment was conducted to modify the standard technique of C-banding, which allowed to visualize C-band of agile gibbons chromosome more clearly and readable for banding analysis.
Although, chromosome painting is an advanced technique of fluorescence in situ hybridization (FISH) and is also very useful for obtaining a high resolution of
more advanced laboratories. The present study was conducted to survey the basic information in chromosomes of small apes that have little been noted so far though actually have informative traits, so that the technique using ordinary facilities were also available. C-bands have little been noted in small apes so far, but actually were useful for detecting a hybrid between different genera (Myers & Shaver 1979; Pellicciari et al. 1988), a unique whole arm translocation within a species of small apes (Hirai et al. 2003), and important traits of chromosomes of agile gibbons that have not been previously uncovered (Wijayanto et al. 2005 in printing). The DAPI staining technique which was used prior to the C-banding in the present study is useful in identifying chromosomes of mammals. Moreover, because DAPI has high affinity to A+T rich DNA, application of DAPI/C-band sequential staining technique allows us to clarify the DNA content of the heterochromatic region of chromosomes. This report will describe detailed methods that we adopted here to detect C-bands and the molecular traits in agile gibbons and siamang that have different C-band features from each other.
Materials and methods
Materials
Blood samples of forty-four agile gibbons and one siamang were obtained from Indonesia (Sumatra and Kalimantan); the Primate Research Institute, Kyoto University, Japan, provided four samples; and siamang blood samples were kindly provided by the Japan Monkey Center, Japan. Whole blood samples (0.7-0.8 mL) were collected with a heparinized syringe from individual agile gibbon anaesthetized with ketamine hydrochloride and cultured as described by Hirai et al. (2003).
Methods
DAPI-C band sequential staining
a. DAPI Staining
Stock Solutions: 5 M NaOH, 20xSSC, 99.5% ethanol, and DAPI Stock
Working solutions: 2xSSC pH 12.5 (5 ml 20xSSC + 500 ul 5M NaOH +
44.5 ml dH2O); 70% ethanol (35 ml ethanol absolute + 15 ml dH2O); BI
buffer (50 ml 1M NaHCO3 + 500 ul Igepal + 450 ml dH2O); DAPI + PI
(Dissolve DAPI stock solution and PI in dH2O and make a final
concentration containing 500 ng/ml DAPI 10, and ng/ml PI).
Procedures were done sequentially as follows:
1) denatured slide by placing in 2xSSC pH 12.5 for 4 minutes, 2) dehydrated slides in 70 % and 99.5% ethanol for 5 minutes each, 3) dried at room temperature,
4) immersed in BI buffer for 5 minutes,
5) mounted with 20 µl anti-fade solution contained DAPI and PI and a cover slip.
DAPI bands were observed by using a Zeiss Axiophoto fluorescence microscope and saved into a computer with a CCD camera system. The good chromosome spread locations were recorded. After saving data, the cover slip of the DAPI-stained slide preparation was removed and the anti-fade solution was washed out with running tap water and the slide was immersed in distillated water for 30 minutes (in Hirai 2001, 1 hour). The slide was treated for C-banding, which is a slightly modified version of the standard technique (Sumner 1972).
b. C-banding staining after DAPI (Barium-Saline-Giemsa/BSG) were done with solutions: 0.2 N HCl (10 ml 1 N HCl + 40 ml dH2O); 5% Ba(OH)2 (2.5 g
Ba(OH)2 + 50 ml dH2O); 2xSSC (0.3 M sodium containing 0.03 M
Sequential procedures:
1) removed cover slip of DAPI-stained slides, wash out anti-fade solution with running tap water and immerse the slides in dH2O
for 30 minutes,
2) soaked in 0.2 N HCl for 30 minutes, 3) washed vigorously in running tap water, 4) treated in 5% Ba(OH)2 53oC for 7 minutes,
5) washed vigorously in running tap water, 6) soaked in 2xSSC 53oC for 5 minutes,
7) stained by 4% Giemsa solution for 30 minutes, 8) washed vigorously in tap water,
9) dried the slides in a 37o
C incubator or at room temperature over night,
10)covered using Malinol mounting solution and a cover slip, 11)observed chromosome spreads, which have had recorded in the
DAPI stained preparation, being able to exactly compare the data of both DAPI-staining and C-banding analyses.
Results and discussion
DAPI staining
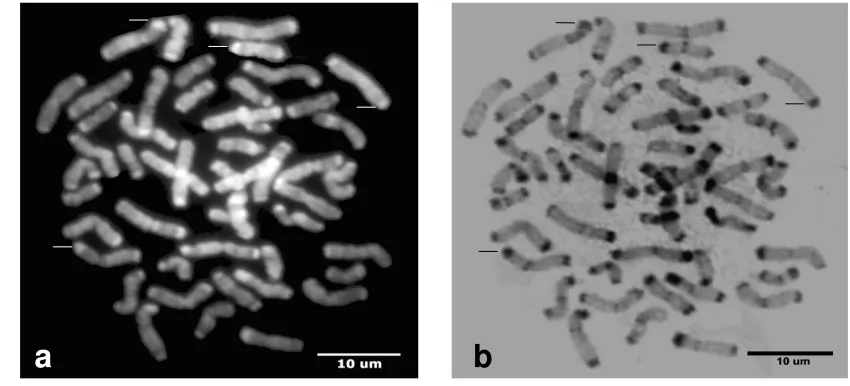
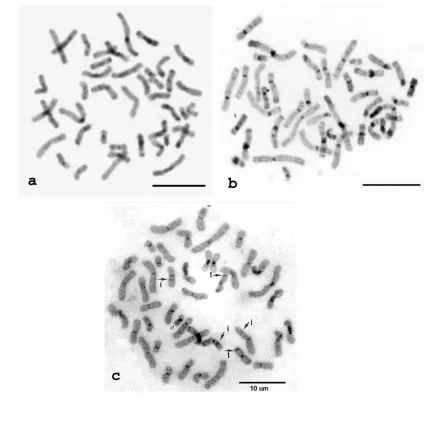
Figure 1. Chromosomes of agile gibbons. DAPI staining (a) and C-band after DAPI-staining (b). Negative DAPI band were expressed as positive C-band (arrowhead) and vice versa (arrow). Scale bar 10 µm
Figure 2. Chromosomes of siamangs. DAPI staining (a) and C-band after DAPI staining (b). Positive DAPI band were also expressed as positive C-band (arrowhead) and vice versa (arrow)
Comparison between DAPI- and C-bands
The standard protocol for BSG (Barium/Saline/Giemsa) C-band staining (e.g., Sumner 1972) was not suitable for staining C-heterochromatin of chromosomes of agile gibbon, though those of siamang showed good results by the standard technique.
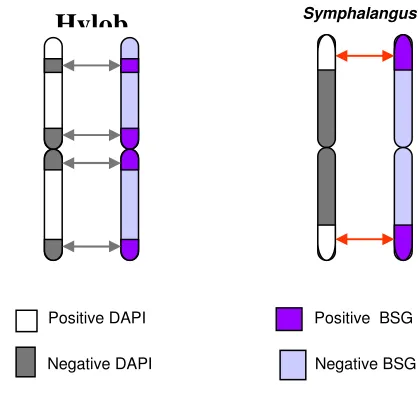
Figure 3. Ideogram explains the expression of DAPI and C-band sequential staining in chromosomes of agile gibbons (Hylobates) (left) and siamangs (Symphalangus) (right). DAPI and C-band between them (arrow) showed opposite traits. Hylobates has C-bands of negative DAPI, while Symphalangus has C-bands of positive DAPI
Several techniques of C-banding were tried to find an adequate procedure for detecting C-bands of agile gibbons that seem to be more vague and complicated rather than other primate species. For example, human chromosomes have C-bands (Figure 4b) that can be detected by a standard technique of BSG established by Sumner (1972), and siamang used in the present species also have the same features of C-band as human, being stainable by the standard technique (see Figure 2b). Finally, the present trials uncovered a best technique using treatment of Ba(OH)2 at
53oC for 7 minutes. Figure 4c shows the best results observed with the modified technique.
Symphalangus
Positive DAPI
Negative DAPI
Positive BSG
Negative BSG
Discussion
The DAPI (4’-6-diamidino-2-phenylindole) staining performed prior to C-banding gave a significant contribution to determining the location of heterochromatic band in the agile gibbon chromosomes, and to detect molecular feature of the bands. DAPI fluorochrome has higher affinity to A+T than to G+C rich DNA (Verma and Babu 1995; Sumner 1990). Therefore, in the present study DAPI/C-band sequential staining was applied to investigate molecular features of C-bands of agile gibbons and siamang. Consequently, C-bands of agile gibbons all accorded with dull (negative) bands of DAPI-staining, meaning that the bands were G+C-rich. On the other hand, terminal C-bands of siamang corresponded with bright (positive) bands of DAPI-staining, meaning that the bands were A+T-rich.
Using the procedure mentioned above, it was found that C-bands of agile gibbons contained G-C rich, while those of siamang had A-T rich DNA. Moreover, the banding analysis showed the C-band pattern of agile gibbons is specific for each chromosome and thus very useful for identification of chromosomes. If the original standard technique of BSG method were used here, C-bands of agile gibbons would have remained unreadable as shown in Figure 4a. As the standard method, BSG method used Ba(OH)2 treatment (Sumner 1972) instead of NaOH to denature DNA
became a good method for obtaining C-band, since it provided an advantage which facilitates better control of the denaturation process than NaOH (Verma and Babu 1995).
Ba(OH)2 is a relatively milder alkali than NaOH, slide preparations must be subjected to a longer treatment with Ba(OH)2 to denature chromosomal DNA, and thus the denaturation process is easier than others. Nevertheless, temperature and time of treatment in Ba(OH)2 is critical to obtain higher quality C-bands. Generally,
characteristics of C-bands of agile gibbons, though the precise mechanism is unknown. That is, lower temperature (from 55°C to 53°C) and shorter time (7 minutes and 25 minutes, respectively) in 5% Ba(OH)2 and 2xSSC treatment were resulted better chromosome morphology and band quality (Figure 4c), although still not as good as human chromosome (Figure 4b).
Figure5. Schematic diagram of C-banding pattern of Hylobates chromosomes (left) with interstitial, terminal, and paracentric heterochromatic bands and Symphalangus (right) with bigger bands at the distal end of biarmed chromosome. Arabic numbers below are chromosome number. Chromosome 12 and 21 of siamang has unique C-bands patterns. As shown, chromosome 12 has telomeric in the short arm and interstitial C-band in the long arm, and chromosome 21 has negative short arm with a centromeric and a telomeric C-bands in the long arm
Treatment temperature 53°C also showed more stable stained ability for C-banding after DAPI staining rather than 55°C used for direct C-banding. This could
Reaction solution Modification method
(in the present study)
Standard method
30 minutes at room temperature 53 oC, 7 minutes
53 oC, 25 minutes
4% Giemsa in Sorenssen’s buffer pH 6.8, 30 minutes
1 hr at room temperature 50 oC, 5-15 minutes 60 oC, 1 hr
2% Gurr’s Giemsa in buffer pH 6.8, 90 minutes
Table 1. Comparation between modified C-band and the standard staining method of Sumner (1972) used in the present study
be related to use of 2.5 pH NaOH 2xSSC solution for DAPI-staining. As with other C band staining methods, freshly prepared slides are usually unsuitable for C-banding, because the chromosome may be too sensitive to withstand harsh treatment, and they thus lose their morphology, becoming fuzzy and hollow in appearance. That is the reason, why the slide preparations have to be aged for either one week or ten days at room temperature or for 2 to 3 days at 50 to 60°C (Verma and Babu 1995). Slide preparations were kept in a 37°C incubator for four to seven days.
Conclusion
Many methods of C-banding have been developed during the last two decades, initially by Sumner (1972), which now becoming a standard method for C-band staining in plant and animal specimen. However, to optimize the method for specific species, researchers in their own laboratory must be doing a specific modification of the technique in order to result in good and readable banding. C-banding which so far have little been noted, was very useful to identify a whole arm translocation in small apes. Amid many very advanced techniques for chromosome analysis, C-banding still become a very important technique for cytogenetic analysis, because such method is very simple, cheap, and applicable for small laboratory in developing countries, which usually have only limited facilities.
Acknowledgement
References
Gustashaw KM (1991) Chromosome Stains. In The ACT Cytogenetics Laboratory Manual, Second Edition, edited by M. J. Barch. The Association of
Cytogenetic Technologists, New York, Raven Press, Ltd.
Hirai H (2001) Relationship of telomere sequence and constitutive heterochromatin in the human and apes as detected by PRINS. Meth Cell Sci 23: 29-35. Hirai H, Mootnick AR, Takenaka O, Suryobroto B, Mouri T, Kamanaka Y, Katoh
A, Kimura N, Katoh A, and Maeda N (2003) Genetics mechanism and property of a whole-arm translocation (WAT) between chromosomes 8 and 9 of agile gibbons (Hylobates agilis). Chromosome Res 11: 37-50.
Hirai H, Wijayanto H, Tanaka H, Mootnick AR, Hayano A, Perwitasari D, Iskandriati D & Sajuthi D (2005) A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features, Chromosome Research 13: 1–11.
Imai HT (1991) Mutability of constitutive heterochromatin (C-bands) during eukaryotic chromosome evolution and their cytological meaning. Jpn J Genet 66: 635-661.
Imai HT, Satta Y, Takahata N (2001) Computer simulations of mammalian and hymenopteran chromosome evolution based on the minimum interaction theory. J theor Biol 210: 475-497.
Myers RH, Shaver DA (1979) Hybrid Ape offspring of mating of gibbon and siamang. Science 205: 308-310.
Pellicciari C, Formenti D, Zuccotti M, Stanyon R, Manfredi Romanini MG (1988) Genome size and constitutive heterochromatin in Hylobates muelleri and Symphalangus syndactylus and in their viable hybrid. Cytogenet Cell Genet 47: 1-4.
Sumner AT (1972) A simple technique for demonstrating centromere heterochromatin. Exp Cell Res 75: 304-306.
Sumner AT (2003) Chromosomes: organization and function. Oxford: Blackwell Scientific.
Verma RS and Babu A (1995) Human Chromosomes, Principles and Techniques, New York. McGraw-Hill, Inc.
Patterns of C-heterochromatin and telomeric DNA in two
representative groups of small apes, the genera Hylobates and
Symphalangus
Hery Wijayanto 1,2,3, Yuriko Hirai1, Yosirou Kamanaka1, Akira Katho4, Sri Supratini Mansjoer2, Dondin Sajuthi2, and Hirohisa Hirai1*
1
Primate Research Institute, Kyoto University, Inuyama, Aichi 484-8506, Japan; Tel:+81-568-630528; e-mail: hirai@pri.kyoto-u.ac.jp; 2
Primate Research Center, Bogor Agricultural University, Bogor Indonesia; 3Department of Veterinary
Anatomy, Gadjah Mada University, Yogyakarta, Indonesia; 4Japan Monkey Center, Inuyama, Aichi, Japan
*
Correspondence
Abstract
Chromosome evolution of small apes is still elusive, though painting analyses have opened the way for elucidating the puzzle. Even C-banding pattern of lar-group gibbons (the genus Hylobates) is not clarified yet, and our previous studies suggested that lar-group gibbons have unique C-banding pattern. We thus conducted to establish C-banded karyotypes of all chromosomes of agile gibbons, a species of Hylobates. The data were compared with those of siamang (the genus Symphalangus) that have distinct C-bands to infer chromosome differentiation of small apes. C-banded chromosomes of agile gibbons showed several terminal, interstitial, and paracentric bands, whose traits are specific for each chromosome, whereas those of siamang were located only at both terminal regions in most chromosomes. Moreover, the C-bands of Hylobates and Symphalangus were emerged to be G+C-rich and A+T-rich DNA, respectively, by DAPI/C-band sequential staining. Furthermore, the PRINS labeling with a telomere primer revealed that agile gibbons have telomeric DNA only at termini where there is no C-band (nontelomeric heterochromatin), whereas telomeric DNA of siamang is located at the same areas at terminal C-banded regions (telomeric heterochromatin). Though the evolutionary mechanism is still unknown, such features seem to be also informative items to infer evolutionary pathway of small apes.
Introduction
Recently, four subgenera, Bunopithecus, Hylobates, Symphalangus, and Nomascus, of the family Hylobatidae (small apes) are all ranked up as genera in a
taxonomist group (Brandon-Jones et al. 2004), though there are still some controversies. Chromosome number is a good landmark to distinguish the genera of small apes, that is, Bunopithecus (2n = 38), Hylobates (2n = 44), Symphalangus (2n = 50), and Nomascus (2n = 52). However, the chromosome evolution contains also an unsolved problem distinct from other primate groups, because small apes might have evolved through extreme number of translocations (e.g., Jauch et al. 1992; Müller et al. 2003). More recently, a molecular analysis showed that the genetic distances among the four subgenera are in the same range as those between the genera Homo and Pan, suggesting to rank as genus, and revealed that the evolutionary branching sequence was regarded as an order of Nomascus [Symphalangus [Bunopithecus and Hylobates]] (Ross & Geissmann 2001).
However, a molecular cytogenetic analysis suggested that the genus Bunopithecus is the most basal group of Hylobatidae, followed by Hylobates, with Symphalangus and Nomascus as the last to diverge (Müller et al. 2003). Furthermore, it has long been demonstrated that the tempo of evolutionary change differs among data sets of phenotypes and genomic markers, respectively.
Furthermore, because, even if heterochromatin is genetically inactive chromatin, it has important several functions, e.g., specific genes, genetic factors, chromosome segregation, and position effect variegation (reviewed in Sumner 2003). Actually, position effect variegation and chiasma reduction by genomic wasteland containing heterochromatin (RCRO) were suggested in chimpanzees (Hirai et al. 2005).
In the recent observations, it was uncovered that lar-group gibbons have more complicated C-banding pattern rather than only-centromeric C-bands described in previous studies, although those are not yet classified into each chromosome (Hirai et al. 2003). The present study depicts the detail C-banded karyotypes of agile gibbons as representative of the genus Hylobates, and demonstrates difference of C-banding pattern and the molecular trait between lar-group gibbons and siamang using techniques of DAPI/C-band sequential staining and PRINS localization for telomeric DNA, and discusses evolutionary aspects of C-heterochromatin differentiation among genera of small apes.
Material and methods
Samples
Blood samples of four agile gibbons were obtained from the Primate Research Institute, Kyoto University, Japan and four siamangs, from Japan Monkey Center, Japan. Whole blood samples (0.7-0.8 mL) collected with a heparinized syringe from individual small apes, anaesthetized with ketamine hydrochloride, were cultured as described previously (Hirai et al. 2003).
DAPI/C-band sequential staining
system. After saving data, the cover slip of the DAPI-stained slide preparation was removed and the anti-fade solution was washed out with running tap water and immersing in distilled water for 1 hr (Hirai 2001). The slide was treated for C-banding, which is a slightly modified version of a standard technique from Sumner (1972). Briefly, the chromosome preparation was treated with 0.2N HCl for 30 min, soaked in 5% barium hydrochloride at 53°οC for 7 min, rinsed in 2xSSC at 53°οC for 25 min, and finally stained with 4% Giemsa (Merck) in Sörenssen’s buffer (pH 6.8) at room temperature for 30 min. The chromosome preparation was completely washed out of each reaction solution under running tap water at the intervals between each step. After drying, the stained slide was covered with a mounting reagent (Malinol) and a cover slip, and the C-banded chromosomes were analyzed for chromosome spreads examined DAPI bands. Treatment temperature 53οC showed more stable stainability for C-banding after DAPI staining rather than 55 oC used for direct C-banding, being also need to get clearer standard C-band patterns.
PRINS localization of telomeric DNA
Telomeric sequences (T2AG3)n were located using PRINS (primed in situ) technique as described by Hirai (2001). Reaction solution was prepared following the manufacturer’s protocol (Boehringer Mannheim, Dig -PRINS reaction kit (no longer products). The results were observed and saved using a Zeiss Axiophoto epifluorescence microscope with a cooled CCD camera system (Photometrics, PXL) connected to a computer (G4, Apple Macintosh), running with IPLab software (Scanalytics).
Nomenclature for classifying C-banded chromosomes