www.elsevier.com/locate/ibmb
Isolation of a cDNA encoding a CHH-family peptide from the
silkworm Bombyx mori
Hirotoshi Endo
a, Hiromichi Nagasawa
b, Toshiki Watanabe
a,*aLaboratory of Molecular Biology of Marine Organisms, Ocean Research Institute, The University of Tokyo, 1-15-1 Minamidai, Nakano, Tokyo
164-8639, Japan
bDepartment of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657,
Japan
Received 30 August 1999; received in revised form 29 November 1999; accepted 13 December 1999
Abstract
The crustacean hyperglycemic hormone (CHH) peptide family includes four types of neuropeptide in decapod and isopod crus-taceans, and the ion-transport peptide in orthopteran insects. To identify a new member of this family in Insecta, a PCR-based search for cDNAs encoding CHH-family peptides was carried out in the silkworm Bombyx mori. A cDNA, named BmCHHL (Bombyx mori CHH-like protein), with an open reading frame of 110 amino acids was isolated. Sequence analyses suggested that the conceptual protein was a precursor of a peptide of 72 amino acids which was amidated at the carboxy terminus. The BmCHHL sequence exhibited significant similarities to members of the CHH family including the orthopteran ion-transport peptide. BmCHHL expression was detected in five or six cells (per hemisphere) in the frontal area of the brain in day 4 fifth instar larvae. 2000 Elsevier Science Ltd. All rights reserved.
Keywords: Silkworm; CHH family; Ion-transport peptide; Neuropeptide; cDNA sequence
1. Introduction
The CHH peptide family comprises peptides that have been isolated from arthropods including crustaceans, orthopteran insects and an arachnid, and is named after the first member of the family, the crustacean hypergly-cemic hormone (Keller, 1992). CHH-family peptides consist of 69–78 amino acids including six conserved cysteine residues which form three intramolecular disul-fide bonds.
The following four neuropeptides of this family have been identified in crustaceans: CHHs, molt-inhibiting hormones (MIHs), vitellogenesis-inhibiting hormones (VIHs) and mandibular organ-inhibiting hormones (MOIHs) (Keller, 1992; Wainwright et al., 1996; Liu et al., 1997) These crustacean peptides are released from the X-organ sinus gland complex which is located in the medulla terminalis of the eyestalk. CHHs regulate the
* Corresponding author. Tel.:+81-3-5351-6534; fax: +81-3-5351-6488.
E-mail address: [email protected] (T. Watanabe).
0965-1748/00/$ - see front matter2000 Elsevier Science Ltd. All rights reserved. PII: S 0 9 6 5 - 1 7 4 8 ( 9 9 ) 0 0 1 2 9 - 0
level of glucose in the hemolymph. MIHs and MOIHs inhibit the release of ecdysteroids from the Y-organ and methyl farnesoate from the mandibular organ, respect-ively, and VIHs inhibit vitellogenesis. All of the above peptides have been identified in decapod crustaceans, except for one CHH isolated from the isopod
Armadillid-ium vulgae (Martin et al., 1993).
In Arachnida, a CHH-family peptide of unknown function was isolated from the venom of the black widow spider Latrodectus mactans tredecimguttatus (Gasparini et al., 1994).
ITP-L has not been elucidated (Macins et al., 1999). cDNAs encoding ITP and ITP-L were also cloned in Locusta
migratoria (Macins et al., 1999).
It is an intriguing question whether insects, like crus-taceans, make use of multiple CHH-family peptides to regulate physiology, development and/or reproduction. It also remains unknown whether the ITP is unique to orthopterans, or common to other insect orders. To address these questions, the authors set out to search for cDNAs encoding CHH-family peptides in the silkworm
Bombyx mori (order Lepidoptera). Here we report the
first isolation of a cDNA encoding a CHH-family pep-tide in non-orthopteran insects.
2. Materials and methods
2.1. Experimental animals
Fourth instar B. mori larvae were purchased from Kanebo Silk Elegance, Co., and grown at 25°C until sac-rificed during fifth instar.
2.2. PCR amplification of B. mori genomic DNA
B. mori genomic DNA was generously provided by
Dr Haruhiko Fujiwara. The polymerase chain reaction (PCR) reaction with degenerate oligonucleotide primers was carried out as previously reported (Watanabe et al., 1996), except that denatured genomic DNA was added to 1 ng/µl.
2.3. Preparation of RNA
The brain [to which the CC and corpus allatum (CA) were attached] and the suboesophageal, prothoracic and mesothoracic ganglia, were collected from 200 fifth instar larvae. The mesothoracic ganglion was only occasionally included. The mixture of tissues was homo-genized in 6 ml of Isogen (Wako), and total RNA was prepared according to instructions from the manufac-turer. Poly(A)+RNA was prepared using Oligotex-dT30 super (Roche Japan).
2.4. 59 Rapid amplification of cDNA ends (RACE)
To 100 ng of the total RNA, 100 pmol of primer CHHR (Fig. 1) was annealed, and cDNA was synthe-sized and poly(A)-tailed. The cDNA was used as a tem-plate in a PCR reaction with the following three primers: a 39 primer (59-GCAGATTCTGTCGAGACG-39), and two 59 primers (59-GAGTCGACTCGAGAATTCT17-39
and 59-GAGTCGACTCGAGAATTC-39). The product
of this reaction was subcloned in pCR2.1 (Invitrogen), and analyzed for the nucleotide sequence.
2.5. Construction and screening of a cDNA library
cDNA was synthesized with 1µg of the poly(A)+
RNA using a Time Saver cDNA synthesis kit
(Pharmacia Biotech), and ligated to the λZAPII vector (Stratagene). The ligation product was packaged using Gigapack Gold Packaging Extract (Stratagene).
The 59 RACE product was radio-labeled to be used as a probe to screen the library. Procedures for library screening have previously been described (Watanabe et al., 1996).
2.6. Nucleotide sequence analysis
Nucleotide sequences of cDNA clones were determ-ined, and conceptual translation performed as has been described (Watanabe et al., 1996). In order to reveal similarities between the BmCHHL protein sequence and previously reported protein sequences, a homology search was performed in ‘Protein All’ (version 3.0t84) databases (including PIR and SWISS-PROT) of DNA Databank of Japan using the FASTA program (Pearson and Lipman, 1988).
2.7. Northern hybridization and reverse transcription (RT)-PCR
Poly(A)+ RNA (0.5µg) prepared as described above was electrophoresed and blotted on to a nylon mem-brane, and hybridization was performed as previously described (Watanabe et al., 1996). A HindIII fragment of 515 bp [A622–A1137nucleotide positions based on Fig. 2(A)] was used as the hybridization probe. After hybridization the filter was washed under a low strin-gency condition (2×SSPE, 37°C).
For RT-PCR analysis, total RNA was isolated from staged fifth instar larvae. An anti-sense probe, the sequence of which was inverse-complementary to A247– C264 [nucleotide positions based on Fig. 2(A)], was annealed to the total RNA samples and cDNA was syn-thesized as previously described (Watanabe et al., 1996). The cDNA samples were used as templates in a PCR reaction (24 or 27 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s) using the following two primers: the 59 primer corresponding to C75–A95[Fig. 2(A)] and the 39 primer inverse-complementary to A247–C264. PCR products were run on 6% polyacrylamide gels, and detected with ethidium bromide. A control experiment in which the reverse transcriptase step was omitted was also performed.
2.8. In situ hybridization
sense riboprobes were generated with T7 and T3 RNA polymerases (PROMEGA), respectively. The latter probe was used as a negative control. The whole mount in situ hybridization was performed on the brain (with attached CC and CA), and suboesophageal and protho-racic ganglia, from day 4 fifth instar larvae as previously reported (Tsuzuki et al., 1997), except that non-fat dry milk was used as the blocking agent instead of sheep serum and that the methyl salicylate clarification step was omitted.
3. Results
3.1. Isolation of a cDNA encoding a CHH-family peptide
As the first step in isolating a cDNA encoding a CHH-family peptide, degenerate oligonucleotide PCR primers were designed based on amino acid sequences of regions conserved among CHH-family peptides [Fig. 1(A)]. The amino acid sequences of six CHH-family peptides were aligned to find highly conserved regions. Two regions (Cys7
–Asp12
and Cys23 –Asn28
in the locust ITP) were thereby selected, and two degenerate oligonucleotide pri-mers (CHHF and CHHR) were designed by reverse translation [Fig. 1(B)].
Genomic DNA prepared from B. mori was used as the template in a PCR reaction using CHHF and CHHR. Multiple bands were observed when the PCR product was separated by polyacrylamide gel electrophoresis and detected with ethidium bromide (data not shown). A band of about 65 bp, the size expected for a CHH-family peptide, was excised from the gel, and DNA eluted from the gel was subjected to another round of PCR amplifi-cation. The product of the second PCR was subcloned and analyzed for the nucleotide sequence. cDNA pre-pared from the brain (with CC and CA attached), and the suboesophageal, prothoracic and mesothoracic gang-lia, was also used as the PCR template, but a product of the expected size was not detected.
One of the subclones analyzed contained a PCR pro-duct of 65 bp, including 34 bp derived from the two pri-mers [Fig. 1(C)]. Its amino acid sequence, when concep-tually translated in one of the reading frames, exhibited significant similarities to CHH-family peptides such as the ITP of the locust, S. gregaria (Meredith et al., 1996), and a CHH of the crayfish, Orconectes limosus (Kegel et al., 1991) [Fig. 1(C)]. Thus, the PCR product was likely to be derived from mRNA encoding a CHH-fam-ily peptide.
Next, the nucleotide sequence (253 bp) of a 59portion
of the cDNA was obtained by 59 RACE (data not
shown), and this RACE product was used as a probe to screen a cDNA library (about 200,000 clones) of the brain (with attached CC and CA), and the
suboeso-phageal, prothoracic and mesothoracic ganglia. Two positive clones were isolated. One of them [1796 bp excluding the putative poly(A) tail] was analyzed for the nucleotide sequence [Fig. 2(A)]. The clone contained a sequence of 31 bp [T231–C261 in Fig. 2(A)] that was identical to the PCR product, and another [C1–C236 in Fig. 2(A)] identical to a 39 part of the RACE product (data not shown). As described below, this cDNA con-tained an open reading frame (ORF) for a CHH-family peptide.
3.2. Inferred structure of the encoded protein
In the clone analyzed, an ORF of 110 amino acids was found near the 59 end [Fig. 2(A)]. This conceptual protein was named BmCHHL due to its sequence larities to CHH-famaily peptides. In order to reveal simi-larities between the conceptual BmCHHL sequence and previously reported protein sequences, a computer-aided homology search was performed. Five proteins with the highest similarity score were the ITP and ITP-L of the locusts S. gregaria and L. migratoria (Meredith et al., 1996; Macins et al., 1999), and the MOIH of the spider crab Libinia emarginata (Liu et al., 1997).
A computer-aided analysis (Nielsen et al., 1997) sug-gests that the conceptual BmCHHL protein contains a signal peptide with the likeliest signal cleavage site between Ala23 and Leu24. The protein is likely to be further processed between Arg35 and Ser36, since this putative processing site is preceded by a dibasic sequence (Arg34–Arg35), and the ITP precursor of S. gre-garia is processed at the corresponding site (Meredith et
al., 1996). The three amino acids at the C-terminus of the ORF (Gly108–Lys109–Arg110) are likely to constitute the site of cleavage and amidation, as many CHH-family peptides are amidated at the C-terminus (Keller, 1992). Based on these results, we presume that the concep-tual BmCHHL protein is a prepropeptide consisting of a signal peptide (Met1–Ala23), a decapeptide (Leu24– Glu33
), a dibasic cleavage site (Arg34 –Arg35
), a peptide of 72 amino acids (Ser36–Val107), and an amidation sig-nal (Gly108–Arg110).
Fig. 1. PCR amplification of a fragment of a cDNA encoding a CHH-family peptide in B. mori. (A) Designing PCR primers. Alignments of the following CHH-family peptides are shown: a crab (Carcinus maenas) CHH (Kegel et al., 1989), a crayfish (Orconectes limosus) CHH (Kegel et al., 1991), a lobster (Homarus americanus) CHH (Chang et al., 1990), a woodlice (Armadillium vulgae) CHH (Martin et al., 1993), a prawn (Penaeus japonicus) CHH (Yang et al., 1995; Ohira et al., 1997) and a locust (Schistocerca gregaria) ITP (Meredith et al., 1996). These peptides belong to ‘Type I’ subgroup of the CHH family (see Discussion). Amino acids that are identical to those in the ITP sequence are indicated by dots, and conserved cysteine residues by asterisks. The two PCR primers (CHHF and CHHR) are derived from highly conserved regions (underlined). (B) Nucleotide sequences of the two degenerate oligonucleotide primers. (C) Nucleotide sequence of a PCR product using B. mori genomic DNA as the template. Sequences corresponding to the primers are underlined. Conceptual amino acid sequence in one reading frame is shown in the one-letter representation above the respective codons. The derived sequence of 10 amino acids is aligned against the corresponding regions of the locust ITP and the crayfish CHH. The amino acids that are identical to those in the B. mori sequence are indicated by dots.
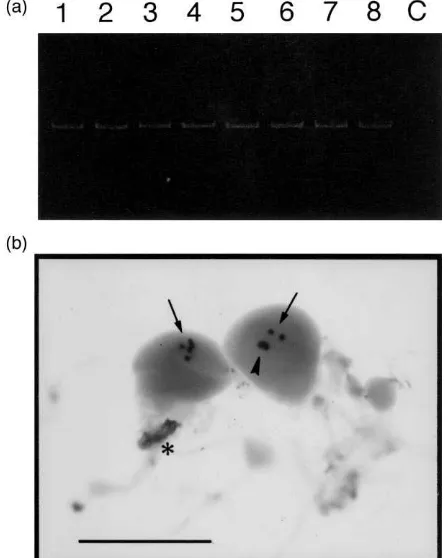
3.3. Expression of BmCHHL
BmCHHL transcripts were detected using RT-PCR in
total RNA samples prepared from the brain (with CC and CA attached), and the suboesophageal, prothoracic and mesothoracic ganglia, of fifth instar larvae. After 27 cycles of amplification, PCR products were clearly detected, and no significant change in the level of BmCHHL was observed during the first eight days of the fifth larval instar [Fig. 3(a)]. When the cycle was reduced to 24, the signals observed were much weaker and barely above the detection level, and even then no significant difference in the signal level was seen among staged samples (data not shown). BmCHHL transcripts could not be detected in Northern analysis, presumably due to the small number of cells expressing BmCHHL. Cells expressing BmCHHL were identified by whole mount in situ hybridization on the brain (with CA and CC attached), and the suboesophageal and prothoracic
ganglia, dissected from day 4 fifth instar larvae (N=2).
BmCHHL expression could be detected only in a small
number of cells in the frontal area of the brain. The num-ber of cells expressing BmCHHL was five per brain hemisphere in one individual [Fig. 3(b)] and six in the other (data not shown). In a control experiment with a sense probe, these cells were not stained (data not shown). Strong signals were also found in tracheal debris attached to the brain and the ganglia. They are not due to BmCHHL expression, but to non-specific hybridiz-ation of the probe, because similar signals were detected in a control experiment using the sense probe (data not shown).
4. Discussion
Fig. 2. Nucleotide sequence of the cDNA clone and deduced amino acid sequence of BmCHHL. (A) Nucleotide sequence of the cDNA clone and conceptually translated sequence of an open reading frame of 110 amino acids. The amino acid sequence is shown in the one-letter representation below the respective codons. The positions of the nucleotides and amino acids are indicated on the left of the sequences. The nucleotide sequence identical to the PCR product (T231–C261) is underlined. ‘u’ indicates the presumptive signal cleavage site (between Ala23and Leu24). The putative
dibasic cleavage site (Arg34–Arg35) and amidation signal (Gly108–Arg110) are indicated in shadowed fonts, and the putative mature BmCHHL peptide
in bold fonts (Ser36–Val107). An asterisk indicates a termination codon. A sequence (A1776–A1781) similar to the consensus polyadenylation signal
(AATAAA) is underlined. The accession number of this sequence in the DDBJ/EMBL/GenBank nucleotide sequence databases is AB031074. (B) FASTA alignment of the Schistocerca gregaria ITP (the upper sequence) and the putative mature BmCHHL peptide (lower). The numbers on the left of the sequences indicate positions of amino acids in the ORFs. An asterisk indicates amino acid identity between the two sequences, and the six conserved cysteine residues are underlined.
higher sequence similarities to the orthopteran ITPs and ITP-Ls than the crustacean peptides of the CHH family. This observation suggests that BmCHHL is a lepidop-teran ortholog of ITP or ITP-L. BmCHHL is more likely to be an ortholog of the former, because the locust ITP precursors, like the BmCHHL precursors, contain puta-tive amidation signals at the C-termini, whereas the
ITP-L precursors do not (Meredith et al., 1996; Macins et al., 1999). Thus, the sequence analysis suggests the possibility that BmCHHL is a lepidopteran ITP.
Fig. 3. Expression of BmCHHL transcripts. (a) Amplification of
BmCHHL transcripts by RT-PCR (27 cycles). A product of 190 bp
(arrowhead) was detected in RNA samples from staged fifth instar lar-vae [days 0–7 (lanes 1–8) after ecdysis], but not in a control sample (lane C). (b) Identification of BmCHHL-expressing cells by whole mount in situ hybridization. Expression was detected in five cells (per hemisphere) in the frontal area of the brain (arrows) from a fifth instar larva. In the hemisphere shown on the right, three expressing cells form a tight cluster (arrowhead). Non-specific hybridization of the probe was seen to tracheal debris (asterisk). Scale bar, 500µm.
may not be universally applicable in Insecta, because a search carried out in the fruitfly Drosophila
melanogas-ter (order Dipmelanogas-tera) was not successful.
CHH-family peptides in Crustacea are grouped into two subgroups (Types I and II) based on the primary structure of the mature peptide and the precursor (De Kleijn and Van Herp, 1995; Yang et al., 1995). Type II peptides have an insertion of a Gly residue near the N-terminus, and the precursors consist only of a signal pep-tide and a mature peppep-tide. By contrast, the precursors of Type I peptides comprise a signal peptide, a peptide of
unknown function (called CHH precursor-related
peptide) followed by a dibasic cleavage site, and the mature hormone, which is often followed by an amid-ation signal. BmCHHL belongs to Type I, since it lacks the Gly insertion, and its precursor structure resembles that of Type I peptides. The locust ITP and ITP-L also belong to Type I, based on the lack of the Gly insertion and presence of a dibasic cleavage site preceding the mature peptide, although the precursors do not contain an apparent signal sequence.
In the present study we designed a set of primers for cDNAs encoding Type II peptides, and conducted searches in B. mori and D. melanogaster, but no such cDNAs could be isolated. Thus, divergence of the CHH family into the two subgroups has been observed only in Crustacea.
The ITP in S. gregaria is synthesized in the brain and released from the CC (Meredith et al., 1996; Macins et al., 1999). In the present study, expression of BmCHHL was also detected in the brain of B. mori larvae. Com-parison of expression patterns of BmCHHL and the locust ITP in the brain is not currently feasible, because in situ hybridization or immunodetection analysis to identify cells expressing the ITPs has not been reported.
BmCHHL expression was detected in a small number
(five or six per hemisphere) of cells in the frontal region of the brain. The difference in the number of expressing cells may be due to a slight difference in the develop-mental stage. In the brain of B. mori larvae, neurosecre-tory cells expressing peptide hormones such as
protho-racicotropic hormone (Kawakami et al., 1990),
bombyxin (Mizoguchi et al., 1987; Iwami, 1990) and eclosion hormone (Kono et al., 1990; Kamito et al., 1992) have been identified. The locations of these cells in the brain are distinct from that of the BmCHHL expressing cells.
Acknowledgements
The authors are grateful to Dr Haruhiko Fujiwara for provision of B. mori genomic DNA, and Dr Yoshiaki Tanaka for provision of B. mori larvae. This work was in part supported by a Grant-in-Aid for Scientific Research (No. 08276204) to TW from the Ministry of Education, Science, Sports and Culture of Japan.
References
Audsley, N., Macintosh, C., Phillips, J.E., 1992. Isolation of a neuro-peptide from locust corpus cardiacum which influences ileal trans-port. The Journal of Experimental Biology 173, 261–274. Chang, E.S., Prestwich, G.D., Bruce, M.J., 1990. Amino acid sequence
of a peptide with both molt-inhibiting and hyperglycemic activities in the lobster Homarus americanus. Biochemical and Biophysical Research Communications 171, 818–826.
De Kleijn, D.P.V., Van Herp, F., 1995. Molecular biology of neurohor-mone precursors in the eyestalk of Crustacea. Comparative Bio-chemistry and Physiology 112B, 573–579.
Gasparini, S., Kiyatkin, N., Drevet, P., Boulain, J.-C., Tacnet, F., Ripo-che, P., Forest, E., Grishin, E., Menez, A., 1994. The low molecular weight protein which co-purifies with a-latrotoxin is structurally related to crustacean hyperglycemic hormones. The Journal of Bio-logical Chemistry 269, 19803–19809.
Kamito, T., Tanaka, H., Sato, B., Nagasawa, H., Suzuki, A., 1992. Nucleotide sequence of cDNA for the eclosion hormone of the silk-worm, Bombyx mori, and the expression in a brain. Biochemical and Biophysical Research Communications 182, 514–519. Kawakami, A., Kataoka, H., Oka, T., Mizoguchi, A.,
Kimura-Kawak-ami, M., Adachi, T., IwKimura-Kawak-ami, M., Nagasawa, H., Suzuki, A., Ishi-zaki, H., 1990. Molecular cloning of the Bombyx mori protho-racicotropic hormone. Science 247, 1333–1335.
Kegel, G., Reichwein, B., Weese, S., Gaus, G., Peter-Katalinic, J., Keller, R., 1989. Amino acid sequence of the crustacean hypergly-cemic hormone (CHH) from the shore crab, Carcinus maenas. FEBS Letters 255, 10–14.
Kegel, G., Reichwein, B., Tensen, C.P., Keller, R., 1991. Amino acid sequence of crustacean hyperglycemic hormone (CHH) from the crayfish, Orconectes limosus: emergence of a novel peptide family. Peptides 12, 909–913.
Keller, R., 1992. Crustacean neuropeptides: structures, functions and comparative aspects. Experientia 48, 439–448.
Kono, T., Mizoguchi, A., Nagasawa, H., Ishizaki, H., Fugo, H., Suzuki, A., 1990. A monoclonal antibody against a synthetic carboxy-ter-minal fragment of the eclosion hormone of the silkworm, Bombyx
mori: characterization and application to immunohistochemistry
and affinity chromatography. Zoological Science 4, 47–54. Liu, L., Laufer, H., Wang, Y., Hayes, T., 1997. A neurohormone
reg-ulating both methyl farnesoate synthesis and glucose metabolism in a crustacean. Biochemistry and Biophysics Research Communi-cations 237, 694–701.
Macins, A., Meredith, J., Zhao, Y., Brock, H.W., Phillips, J.E., 1999. Occurrence of ion transport peptide (ITP) and ion transport-like (ITP-L) in orthopteroids. Archives of Insect Biochemistry and Physiology 40, 107–118.
Martin, G., Sorokine, O., Van Dorsselaer, A., 1993. Isolation and mol-ecular characterization of a hyperglycemic neuropeptide from the sinus gland of the terrestrial isopod Armadillidium vulgare (Crustacea). European Journal of Biochemistry 211, 601–607. Meredith, J., Ring, M., Macins, A., Marschall, J., Cheng, N.N.,
Theil-mann, D., Brock, H.W., Phillips, J.E., 1996. Locust ion transport peptide (ITP): primary structure, cDNA and expression in a baculo-virus system. The Journal of Experimental Biology 199, 1053– 1061.
Mizoguchi, A., Ishizaki, H., Nagasawa, H., Kataoka, H., Isogai, A., Tamura, S., Suzuki, A., Fujino, M., Kitada, C., 1987. A monoclonal antibody against a synthetic fragment of bombyxin (4K-protho-racicotropic hormone) from the silkmoth, Bombyx mori: charac-terization and immunohistochemistry. Molecular and Cellular Endocrinology 51, 227–235.
Nielsen, H., Engelbrecht, J., Brunak, S., Von Heijne, G., 1997. Identi-fication of prokaryotic and eukaryotic signal peptides and predic-tion of their cleavage sites. Protein Engineering 10, 1–6. Ohira, T., Watanabe, T., Nagasawa, H., Aida, K., 1997. Cloning and
sequence analysis of a cDNA encoding a crustacean hyperglycemic hormone from the kuruma prawn Penaeus japonicus. Molecular Marine Biology and Biotechnology 6, 59–63.
Pearson, W.R., Lipman, D.J., 1988. Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences, USA 85, 2444–2448.
Tsuzuki, S., Masuta, T., Furuno, M., Sakurai, S., Iwami, M., 1997. Structure and expression of bombyxin E1 gene: a novel family gene that encodes bombyxin-IV, an insect insulin-related neurosecretory peptide. Comparative Biochemistry and Physiology 117B, 409– 416.
Wainwright, G., Webster, S.G., Wilkinson, M.C., Chung, J.S., Rees, H.H., 1996. Structure and significance of mandibular organ-inhibiting hormone in the crab, Cancer pagurus. The Journal of Biological Chemistry 271, 12749–12754.
Watanabe, T., Kono, M., Aida, K., Nagasawa, H., 1996. Isolation of a cDNA encoding a putative chitinase precursor in the Kuruma prawn Penaeus japonicus. Molecular Marine Biology and Biotech-nology 5, 299–303.