The rearranged genetic code and its implications in
evolution and biochemistry
Yuhuang Qiu
a, Lihuang Zhu
b,*
aInstitute of Automation,Academia Sinica,Beijing 100080, PR China bInstitute of Genetics,Academia Sinica,Beijing 100101, PR China
Received 22 October 1999; received in revised form 22 March 2000; accepted 29 March 2000
Abstract
We rearrange the genetic code and present a table of codons. The chemical properties of the amino acids coded by codons, and the evolutionary trend of codons are well reflected in the order of this table, from which two rules can be drawn: (1) the polarity/non-polarity and hydrophilicity/hydrophobicity of amino acids coded for by codons alternate row by row in the table; (2) in general, the lower down in the table, the earlier the codons are in terms of evolution. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Genetic code; Evolution; Biochemistry; Codons
www.elsevier.com/locate/biosystems
Since the genetic code was disclosed, many researchers have tried direct numerical evaluation of the genetic code in order to explain what content the codons may involve. Alvager et al. (1989) worked on the degeneracy of codons based on information theory. Shcherbak (1993) thought that a certain formalization is possible on the level of molecular structures of amino acids though they are a more complicated object for formal procedures than triplets. Swanson (1984) arranged codons in a codon ring, brought codons in corre-spondence with Gray codes, and proposed a uni-fying concept for the amino acid code. On the basis of Swanson codes, Rakocevic (1998)
rear-ranged codons and presented that the Golden mean is a key determinant and invariant of the genetic code. In this paper we rearrange codons and present a new table for the genetic code, which is more systematic than others. To some extent this table can reflect the evolutionary trend of the codons and the relationship between the codon compositions and the properties of the encoded amino acids.
1. The arrangement of the codons
In the Watson – Crick model, DNA forms a double helix which is held together by hydrogen bonds between specific pairs of bases, adenine (A) to thymine (T) and guanine (G) to cytosine (C). * Corresponding author. Fax: +86-10-64873428.
E-mail address:lhzhu@igtp.ac.cn (L. Zhu)
We represent A – T and G – C bonding by ‘0’ and ‘1’. There are two hydrogen bonds between A and T and three between G and C, so we use two and three binary digits to denote A – T and G – C, respectively.
Let
A=10 G=111 T=01 C=000
In messenger RNA thymine (T) is substituted by uracil (U), so U=01.
In order to denote codons by the number series of the quaternary system, we do modulo-opera-tion (mod). Because the residue for 7 modulo-4 is 3, so we have following forms,
A=22 (mod 4) G=73 (mod 4) U=11 (mod 4) C=00 (mod 4)
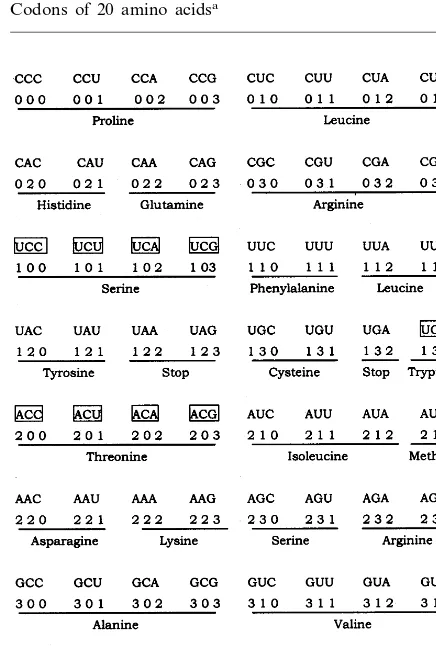
Sixty-four triplets of the universal code consist of a complete set of three-digit numbers of the qua-ternary system. The numbers are arranged as the natural number sequence, 0 – 63. The table of codons is shown in Table 1. For convenience, we formulate the symbol as follows:
(1) R=a puRine (A or G)
(2) Y=a pYrimidine (U or C)
(3) N=a purine or pyrimidine (aNy base)
Table 1 has eight horizontal rows and vertical columns. Each row consists of codons, three-digit numbers of the quaternary system and corre-spondmg amino acids. From up to down, we denote each row with a class symbol, i.e. CYN, CRN, UYN, URN, AYN, ARN, GYN and GRN, respectively.
2. The properties of amino acids
Amino acids can be classified according to their electric charge and the affinity of their side chains for water. The connections between the properties of an amino acid and the mid-base of its codons have been revealed by Taylor and Coates. It is noted that the table of the rearranged genetic code is able to reflect this relationship orderly and periodically. In Table 1 non-polar amino acids are situated in odd rows while polar amino acids are in even rows. However, there are three exceptions to this rule, i.e. serine in the third row, trytophan in the fourth row, and threonine in the fifth row. Nevertheless, the serine encoded by two codons (AGC and AGU) in the sixth row accords with the polarity of the amino acids in this row. In fact, this rule can be expressed in terms of bio-chemistry, i.e. the codons, whose mid-base is a pyrimidine, code non-polar amino acids (except the codons for serine m the third row and the codons for threonine) while the codons, whose mid-base is purine, code polar amino acids (ex-cept the codons for tryptophan).
Table 1
Codons of 20 amino acidsa
aThe codons coding amino acids, that are different in the
There is a similar rule for hydrophobicity/ hy-drophilicity of amino acids. In general, the codons in odd rows and those in even rows code hydro-phobic and hydrophilic amino acids, respectively. And again only serine, tryptophan and threonine in the third, fourth and fifth row, respectively, are irregular. According to Taylor and Coates (1989), the codons whose mid-base is A code for hy-drophilic amino acids, whereas the codons whose mid-base is U code for hydrophobic amino acids. The amino acids, which are coded for by the mid-base G or C codons, are in the middle of the hydrophobicity/hydrophilicity spectrum of amino acids (Taylor and Coates, 1989). However Taylor and Coates (1989) pointed out that hydrophilicity of arginine (whose codons are CGN or AGR with G in middle position) is one of the major excep-tions to the rule proposed by themselves. Never-theless in our table, arginine is situated in the even rows (the second and the sixth row).
The above rules may be related to the direct interactions between amino acids and nucleotides in the template in the early process of genetic codon evolution, that might predate the tRNA – ribosome system. For example, in a proposed hypothetical primitive template uracil had side chains that recognized specific hydrophobic amino acid side chain coordinately with forma-tion of two hydrogen bonds to the peptide back-bone (Chao et al., 1976). It is also worthwhile to mention that the two exceptional amino acids, serine in the third row and threonine in the fifth row in Table 1, both have a hydroxyl group in their side chains, and that their corresponding codons, UCN and ACN, all have C in the middle. According to Niu et al. cytosine can form two hydrogen bonds to the peptide backbone plus a third one to the hydroxyl group of a serine or threonine residue (Niu et al., 1987), so that three hydrogen bonds form readily between cytosine and a serine- or threonine-containing peptide to produce an intimately fitting complementary structure. In this case pairing is especially strong. The supposed interaction between cytosine and serine or threonine may explain the existence of these two exceptions to the general rule.
Tryptophan is a kind of frequently less com-mon amino acid in proteins. It is the amino acid
that has the most side chain atoms in the 20 amino acids. We can not give a logical explana-tion for its excepexplana-tional posiexplana-tion in this table.
3. The evolutionary trend of codons
Since the genetic code was elucidated, there have been two contrary views on whether codons themselves have had an evolutionary process (Vogel, 1998): one of them is that the genetic code used by all creatures is the frozen result due to a historical inchoate accident in the early evolution process. The other view reasons that the code itself is the result of evolution and selection. This hypothesis has been studied in three ways: first the earliest nucleotide sequence was traced; second, the earliest amino acids were proposed, and in the third way the simplest amino acids in the biosyn-thesis pathways were followed. It should be noted again that in our table the order from the bottom up reflects the evolutionary trends of the codons in the three ways.
Crick et al. (1976) first suggested that the RRY triplet was the original code, and Eigen and Schuster (1978) assumed RNY to be original triplet for genetic code. It is a general thought that to the first position in the original triplets (codons) as a purine G occurred earlier than A (Taylor and Coates, 1989).
Table 2
The genetic codes in human mitochondriaa
aBoxes indicate the codons which are different from general
codons.
isoleucine) that all were found in proteins of early life (Cromin and Moore, 1971; Miller and Orgel, 1974; Eigen and Schuster, 1978). It seems to us that the inchoate codons were well-regulated in the very early life. They became complex from single base changes in evolution; additional base changes led to more complex amino acids, so that some exceptional ones came into being when more amino acids were enrolled in proteins. We assume that in Table 1, the codons of the first, second, fifth, and sixth column in the fifth and sixth row could be second earliest. This also fits the earlier comma-less code hypothesis proposed by Shepherd (1981).
Recently, Trifonov proposed that the first codon is GCU and all the others evolve from it (Pennisi, 1998). He also assumed that the earliest changes were one-letter change in GCU, and two-letter changes were all later (Pennisi, 1998). Though Trifonov’s theory is an assumption, GCU corresponds to not only the form RNY but also the form in which the first base is G. Therefore, we conclude that the codons whose first base is G were the earliest codons, and the codons with the form RNY followed.
The genetic code in mitochondria is some way different from the universal genetic code. For example, the genetic code in human mitochondria has following different characteristics (Anderson et al., 1981).
1. UGA is not used for termination, but instead codes for tryptophan.
2. AGA and AGG are not used for arginine, but instead code for termination.
3. The internal methionine has two codons, i.e. AUG and AUA. The start methionine has four codons, i.e. AUN.
According to the same method the codons in human mitochondria can be arranged more or-derly than universal codons (see Table 2), but the chemical periodicity of amino acids and the evolu-tionary trend are all the same in Tables 1 and 2. This shows that the genetic code in mitochondria probably was a later branch of the universal genetic code after the general relations between tRNAs and amino acids had been established. glutamic acid have similar molecular structure,
and chemical properties, only glutamic acid has one more CH2 than aspartic acid. And both are
acidic and negatively charged (Lewin, 1977). In all the amino acids only these two have this property. Finally, as discovered by Taylor and Coates (1989), the GNN coded amino acids are only found at the head of the synthetic pathway of amino acids. Therefore, the codons in the last two rows of Table 1 might be the earliest.
4. Some other regularities of codons
The rearranged table of the genetic code also reflects the roles of tRNAs since it displays the wobble hypothesis (Crick et al., 1976) more clearly. Besides the wobbles that occur at the third position, the most important characteristic of a codon is whether it has a purine or a pyrimidine at the middle base position. In this respect several rules can be further deduced from the Table 1.
1. If A is the mid-base of four contiguous codons in a row, the four codons must divide into two groups, one with R and the other with Y in the third positions, coding for two different amino acids.
2. If C is mid-base of four contiguous codons in a row, the four codons must be grouped into one coding for one amino acid.
3. If the two codons in a row have the first and second base in common but have C and U in the third position, respectively, they both must code for one amino acid.
In addition, the two amino acids, proline and glycine, situated at the top left and the bottom right, respectively, of the rearranged table, are noticeable for their special properties. Proline coded by CCN is an exceptional amino acid in which the nitrogen atom of the amino group is incorporated into a ring, whereas the amino acids coded for by other codons are all a-amino acids
(Clark, 1977). A proline residue disrupts the usual organization of the backbone of a polypeptide, causing a sharp transition in the direction of the chain. That is important when proteins acquire their correct structure (Lewin, 1977). Glycine coded by GGN is the simplest amino acids with the lowest molecular weight. It appears that there might be a main trend from the codons for glycine to the codons for proline in the early evolution of the genetic code predating the tRNA – ribosomal translation system.
5. Discussion
For the quaternary digits, we could have 24 possible choices to assign four digits to four let-ters. In case the assignment A=3, C=2, G=1
and U=0 was chosen, the natural numbers from 0 to 3 would appear as quaternary numbers 000, 001, 002 and 003, corresponding to codons UUU (phenylalanine), UUG (leucine) and UUC (phenylalanine) and UUA (leucine), respectively, but the two codons, UUU and UUC, for pheny-lalanine and the other two codons, UUG and UUA for leucine are separated by each other. So we do not choose this assignment. We have tried all the assignments, of which 20 give similar re-sults as the above example. Of the other four assignments, where A=2, C=1, G=3, U=0 and A=1, C=2, G=0, U=3 both are not as orderly as we present in this paper. Only the assignment A=1, C=3, G=0, U=2 could gen-erate a code table like Table 1 but with inverted direction. We prefer Table 1 since the evolution-ary trend is usually diagramed as a tree upwards. So A=2, C=0, G=3 and U=1 is the best assignment for the quaternary digits. (The rear-ranged genetic code presented in Table 1 is in accordance with some Previous reports on the order in the genetic code (Shepherd, 1981; Niu et al., 1987; Taylor and Coates, 1989; Pennisi, 1998), but this table reflects evolution and properties of codons more clearly than others. We believe that the obvious orderliness within the genetic code and within the codons themselves must have not been the result of chance, but the reflection of the history of the genetic code. The generation and evolution of the genetic code must follow chemi-cal and biochemichemi-cal regulations.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Nos. 69681002 and 69635030).
References
Alvager, T., Graham, G., Hilleke, R., et al., 1989. On the information content of the genetic code. BioSystems 22, 189 – 196.
Chao, H.M., Hazel, B., Black, S., 1976. Binding of uracil perivatives to hydrophobic peptides and sodium dodecyl sulfate. J. Biol. Chem. 251, 6924 – 6928.
B.F.C. Clark, The genetic code. Edward Arnold, 1977. Crick, R.H.C., Brenner, S., Klug, A., et al., 1976. A
specula-tion on the origin of protein synthesis. Origins of Life 7, 389 – 397.
Cromin, J.R., Moore, C.B., 1971. Amino acid analyses of the Murchison, Murray and Allende carbonaceous chondrites. Science 172, 1327 – 1329.
Eigen, M., Schuster, P., 1978. The hypercycle. A principle of natural self-organization, Part C: The realistic hypercycle. Naturwissenschaften 65, 341 – 369.
B. Lewin, Genes VI, Oxford University Press, Oxford, 1977. Miller, S.L., Orgel, L.E., 1974. The Origins of Life on the
Earth. Prentice-Hall, Englewood Cliffs, NJ.
Niu, C.H., Yeh, H.J.C., Black, S., 1987. Hydrogen bonding between cytosine and peptides of threonine or serine: is it
relevant to the origin of the genetic code? Biochem. Bio-phys. Res. Comm. 148, 456 – 462.
Pennisi, E., 1998. The first codon and its descendants. Science 281, 330.
Rakocevic, M.M., 1998. The genetic code as a golden mean determined system. BioSystems 46, 283 – 291.
Shepherd, J.C.W., 1981. Periodic correlations in DNA se-quences and evidence suggesting their evolutionary origin in a comma-less genetic code. J. Molec. Evol. 17, 94 – 102. Shcherbak, V.I., 1993. Twenty canonical amino acids of ge-netic code: the arithmetical regularities. Part I. J. Theoret. Biol. 162, 399 – 401.
Swanson, L., 1984. A unifying concept for amino acid code. Bull. Math. Biol. 46, 187 – 203.
Taylor, F.J.L, Coates, D., 1989. The code within the codons. BioSystems 22, 177 – 187.
Vogel, G., 1998. Track the history of the genetic code. Science 281, 329 – 331.