www.elsevier.com/locate/ibmb
Cloning of a horn fly cDNA, Hi
a
E7, encoding an esterase whose
transcript concentration is elevated in diazinon-resistant flies
Felix D. Guerrero
*USDA–ARS Knipling–Bushland US Livestock Insects Research Lab, 2700 Fredericksburg Road, Kerrville, TX 78028, USA
Received 17 November 1999; received in revised form 18 April 2000; accepted 19 April 2000
Abstract
Reverse transcriptase-polymerase chain reaction (PCR) was used to clone two esterase cDNAs from a diazinon-resistant field population of horn flies that expresses qualitative and quantitative differences in esterases compared with a susceptible population. The open reading frame from one of the esterase cDNAs, HiaE7, exhibits substantial amino-acid identity to an esterase associated
with diazinon resistance in Lucilia cuprina. RNA Northern blots showed that HiaE7 mRNA was more abundant in the
diazinon-resistant population than the susceptible population. DNA copy number analysis did not reveal major differences in HiaE7 gene
copy number between the two populations. The full-length cDNA to HiaE7 was cloned and sequenced, and found to contain all
of the highly conserved sequence elements associated with carboxyl/cholinesterases. The HiaE7 homologs in diazinon-resistant
strains of L. cuprina and Musca domestica have been shown to possess an amino-acid substitution conferring diazinon hydrolytic activity to the esterase enzyme. This amino-acid substitution was not found in diazinon-resistant horn flies examined by allele-specific PCR. Individual flies from the resistant field population were phenotyped as diazinon-resistant or diazinon-susceptible by topical diazinon application bioassays and total RNA isolated and hybridized to HiaE7 probe in ribonuclease protection assays.
HiaE7 transcript was expressed at a five-fold higher level in resistant female individual flies than in susceptible female individuals.
Published by Elsevier Science Ltd.
Keywords: Diazinon resistance; Ribonuclease protection assays; Sequence; Esterases
1. Introduction
The horn fly, Haematobia irritans (L.) (Diptera: Muscidae), is a hematophageous parasite of cattle. There is a long history of reliance upon the use of insecticides for control of this pest and, as a consequence, there have been outbreaks of resistance in various fly populations (Sparks et al., 1985). Resistance to organophosphates (OPs) had been reported as early as 1963 (Burns and Wilson, 1963). Byford et al. (1999) found that horn flies developed moderate levels of resistance to diazinon in both laboratory and field experiments employing con-tinuous insecticide pressure. Carboxylesterases are a major mechanism of OP resistance in mosquitoes, gener-ally causing resistance by sequestration of the OP before it can reach the target site within the insect (Hemingway
* Fax:+1-830-792-0314.
E-mail address: [email protected] (F.D. Guerrero).
0965-1748/00/$ - see front matter. Published by Elsevier Science Ltd. PII: S 0 9 6 5 - 1 7 4 8 ( 0 0 ) 0 0 0 8 8 - 6
and Karunaratne, 1998). Likewise, the OP resistance of other insects exhibiting a metabolic resistance mech-anism to OPs has been shown to be due to the overpro-duction of esterase as in the saw-toothed grain beetle (Conyers et al., 1998) and the German cockroach (Scharf et al., 1997), among others. However, in Lucilia cuprina, a specific mutation in an esterase gene has been found to confer OP resistance (Newcomb et al., 1997a). The mutated gene, LcaE7, contains a Gly137→Asp
substi-tution within the active site of the esterase conferring OP hydrolase activity. In fact, a second mutated LcaE7
allele has been identified in malathion-resistant popu-lations of L. cuprina. Resistant individuals from these populations possess a Trp251
→Leu substitution which
confers malathion carboxylesterase activity leading to malathion resistance (Campbell et al., 1998). Recently, an OP-resistant strain of Musca domestica was found to possess the Gly137
→Asp amino-acid substitution in the
M. domestica ortholog to LcaE7 (Claudianos et al.,
Several horn fly populations were recently discovered in Texas that were resistant to the OP diazinon. Bio-chemical investigations implicated the overexpression of esterases as a mechanism of resistance (Guerrero et al., 1999). Using topical applications of diazinon to pheno-type individual flies as resistant or susceptible, native polyacrylamide gel electrophoresis (PAGE) analysis found that resistant flies were qualitatively and quantitat-ively different from susceptible flies in their esterase activity gel profiles. Some resistant flies had a higher amount of total esterase activity while others appeared to have a total esterase activity similar to the levels found in susceptible flies but possessed unique gel bands of esterase activity. Since OPs are becoming increasingly relied upon for horn fly control, it seemed prudent to study specific mechanisms of OP resistance in order to develop strategies to maximize the effectiveness of OPs and the period of time before resistance becomes wide-spread. This report describes the cloning and sequencing of two horn fly esterase cDNAs, designated HiaE7 and
HiaE8. HiaE7 encodes the horn fly homolog to the L.
cuprina OP-resistance-conferring esterase, LcaE7. The
expression of HiaE7 and HiaE8 mRNA in individual
OP-resistant and OP-susceptible horn flies was assayed to determine if either of the esterase mRNAs was differ-entially expressed in association with OP resistance.
2. Materials and methods
2.1. Horn fly rearing and pesticide resistance assays
OP-susceptible adult horn flies were obtained from a laboratory colony maintained since 1961 at the USDA– ARS Laboratory in Kerrville, TX. Flies were reared as described by Schmidt et al. (1976) except that the cit-rated (15 mM) bovine blood used to feed adults was treated with 250,000 units of oral-grade mycostatin and 0.5 g kanamycin sulfate per liter and the larval rearing medium consists of two parts by weight bovine feces, one part water and one part peanut hull pellets. A field population of diazinon-resistant horn flies at Camp Cooley in Robertson County, TX was sampled and returned live to the laboratory, where a portion were used for topical diazinon application experiments and the remainder frozen and stored at 280°C. In performing the topically applied diazinon experiments, 0.5µl of diazinon dissolved in acetone was applied to the thorax of individual horn flies by using a hand microapplicator (Burkard, Rickmansworth, UK). Fifty individual adult horn flies of mixed ages were treated with 50 or 5 ng of diazinon in acetone. These two diazinon concentrations had previously been found to be suitable for selection of diazinon-resistant and diazinon-susceptible individ-uals from this population (Guerrero et al., 1999). One set of flies was treated with acetone alone as a negative
control. As mortality occurred, time of death and sex were recorded and the fly was frozen for subsequent analyses.
2.2. RNA preparations
Total and poly(A)+RNA were purified with the RNA-gents Total RNA Isolation System and the PolyATtract System (Promega, Madison, WI). RNA concentrations were determined by absorbance spectrophotometry and RNA analyzed by formaldehyde gel electrophoresis and Northern blotting as described in Ausubel et al. (1998).
2.3. Reverse transcriptase-PCR (RT-PCR)
For RT-PCR, reverse transcription of poly(A)+ RNA from both the laboratory OP-susceptible and the Camp Cooley OP-resistant strains was performed on 1µg of heat-denatured poly(A)+RNA at 37°C for 1 h in a reac-tion including 50 mM Tris–HCl (pH=8.3), 75 mM KCl, 10 mM dithiothreitol, 3 mM MgCl2, 0.2 mM of each
dNTP, 2µM of oligo dT18 and 200 units of M-MLV
reverse transcriptase (Life Technologies, Gaithersburg, MD). The RNA template was then hydrolyzed by the addition of 1.5 volumes of a 0.7 M NaOH/40 mM ethy-lenediamine-N,N,N9,N9-tetraacetic acid (EDTA) sol-ution, incubated for 10 min at 65°C, the solution extracted with a 25:24:1 mixture of phenol:chloro-form:isoamyl alcohol and the cDNA recovered by etha-nol precipitation in the presence of Pellet Paint Co-Pre-cipitant (Novagen, Madison, WI). PCR was performed using a 50µl reaction containing 5% of the reverse tran-scription reaction products, 1 nmol of either degenerate primer FG-144 or FG-145, 50 pmol of primer FG-146 (Table 1), 10 mM Tris–HCl (pH=8.3), 50 mM KCl, 0.2 mM each dNTP, 2 mM MgCl2 and 0.5µl of a 1:1
(v/v) mix of AmpliTaq DNA Polymerase (PE Applied Biosystems, Foster City, CA) and TaqStart Antibody (Clontech, Palo Alto, CA). The amplification program
was as described in Hecker and Roux (1996) and used 96°C for 3 min followed by 30 cycles, each consisting of denaturation at 94°C for 1 min, annealing at 65°C for 2 min with a temperature ramp of 20.5°C/cycle and extension at 72°C for 3 min, followed by 10 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 2 min and extension at 72°C for 3 min, and a final exten-sion at 72°C for 7 min.
2.4. Allele-specific PCR (ASPCR)
To prepare genomic DNA, individual frozen adult flies were crushed in 1.5 ml microcentrifuge tubes using a disposable pellet pestle (PGC Scientifics, Gaithersburg, MD). Next, 25µl of buffer [500 mM KCl, 100 mM Tris–HCl (pH=8.3)] was added and the tube contents incubated for 2 min in a boiling water bath. Following a 5 min centrifugation at 15,000g, an aliquot of the supernatant was diluted 1:10 in water. The ASPCR assay thermocycling parameters consisted of an initial denatur-ation at 96°C for 2 min followed by 35 cycles, each con-sisting of denaturation at 94°C for 1 min, annealing at 66°C for 1 min and extension at 72°C for 1 min, and a final extension at 72°C for 7 min. Reactions contained 1µl of the diluted fly DNA, 10 mM Tris–HCl (pH=8.3), 50 mM KCl, 0.05 mM each dNTP, 2 mM MgCl2, and
0.1µl of a 1:1 (v/v) mix of AmpliTaq DNA Polymerase and TaqStart Antibody, and 20 pmol of both primer 158 and one of the allele-specific primers, 203, 204, 206, 207, 208, 209, 210 or FG-211 (Table 1). Amplification products were detected by agarose gel electrophoresis (Ausubel et al., 1998).
2.5. Cloning and sequencing
For subcloning and sequencing experiments, DNA products from the RT-PCR experiments were purified using agarose gel electrophoresis, extracted with the Qiaex Gel Extraction Kit (Qiagen, Inc., Chatsworth, CA) and subcloned into pT7Blue-2 using the Perfectly Blunt Cloning Kit (Novagen, Inc., Madison, WI). DNA was prepared from selected clones using the QIAprep spin miniprep kit and inserts purified by digestion with restriction enzyme, agarose gel electrophoresis and gel extraction. Probes were labeled with 32P-dATP (New
England Nuclear, Boston, MA) using the Strip-EZ DNA Labeling Kit (Ambion Inc., Austin, TX). Autoradio-grams were analyzed by densitometric scanning and quantified by Collage Image Analysis Software (Fotodyne, Inc., Hartland, WI). The full-length HiaE7
cDNA clone was obtained by screening a cDNA library synthesized in the Uni-ZAP XR vector (Stratagene, La Jolla, CA) from poly(A)+ RNA from the Camp Cooley adults. The library was screened with the partial HiaE7
fragment from the RT-PCR experiment using standard procedures (Ausubel et al., 1998) and in vivo excision
performed according to the cDNA library kit manufac-turer’s protocols. Manual sequencing of both DNA strands was done with the T7 Sequenase Version 2.0 DNA Sequencing Kit (Amersham Life Sciences, Inc., Cleveland, OH). The deduced amino-acid sequence alignments were performed with the pam250S scoring matrix of the MacVector Software Ver. 5.0 (Oxford Molecular Group, Oxford, UK).
2.6. Ribonuclease protection assays
Gene expression assays were performed with gene-specific probes synthesized from restriction-enzyme-digested HiaE7 and HiaE8 cDNA clones. T3 RNA
polymerase, the MAXIscript In Vitro Transcription Kit and the HybSpeed RPA Kit (Ambion, Inc., Austin, TX) were used to generate 32P-labeled antisense strand RNA
probes and perform ribonuclease protection assays (RPA). Total RNA was isolated from individual adult horn flies by a miniprep procedure (Verwoerd et al., 1989). Ultraviolet (UV) absorbance values were used to ensure that equal amounts of total RNA from each RNA miniprep were present in the RPA probe hybridization reactions. Human glyceraldehyde 3-phosphate dehydro-genase (GAPDH) was used as a control probe to verify that RNA concentration was approximately constant between samples. GAPDH had previously been shown to have high levels of expression in all life stages of the horn fly (data not shown). The RPA hybridizations used 0.5µg of total RNA and 30,000 cpm each of radiolab-eled GAPDH, HiaE7 and HiaE8 RNA probe in a single
assay. The template sources for the probes were the 396 bp HiaE7 and 326 bp HiaE8 cDNAs isolated during
the degenerate primer RT-PCR experiments and sub-cloned into pT7Blue-2. Molecular biology grade chemi-cals were obtained from GIBCO BRL (Gaithersburg, MD), Sigma Chemical Co. (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA) when available.
3. Results
3.1. Esterase cDNA cloning
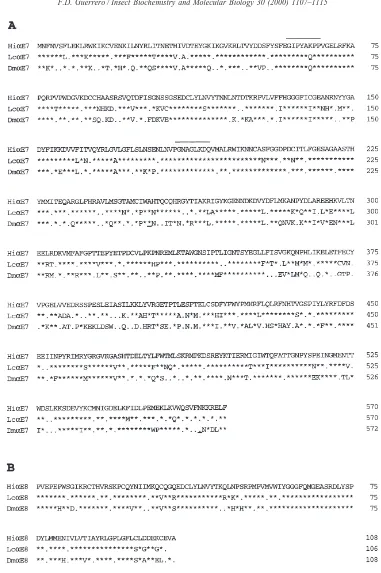
The degenerate primers FG-144, FG-145 and FG-146 were designed from conserved regions of esterase sequences noted by Newcomb et al. (1997b). RT-PCR using FG-144 and FG-146 and poly(A)+RNA from diaz-inon-resistant and diazinon-susceptible flies resulted in the amplification of a 396 bp fragment [Fig. 1(A)] that was found to have substantial DNA sequence homology and amino-acid identity to LcaE7 and DmaE7 from L.
cuprina and Drosophila melanogaster, respectively. In L. cuprina, this region of the LcaE7 gene contains an
amino-acid substitution, Gly137
→Asp, which confers
Fig. 1. Alignment of the deduced amino-acid sequence from the horn fly HiaE7 and HiaE8 cDNAs with previously characterized sequences from L. cuprina and D. melanogaster. The deduced amino-acid sequence of HiαE7 was aligned with LcαE7 and DmαE7 from L. cuprina and D. melanogaster, respectively (A), and HiαE8 was aligned with LcαE8 and DmαE8 from L. cuprina and D. melanogaster, respectively (B). Alignment was performed with the pam250S scoring matrix of the MacVector Software Ver. 5.0. GenBank accession numbers for LcaE7, LcaE8, DmaE7 and DmaE8 are U56636, U49423, U51052 and U51050, respectively. Asterisks (*) indicate an amino-acid identity compared with the HiαE7 or HiαE8 sequences while a dot (.) indicates a conservative amino-acid substitution. An underscored residue (F) indicates that an amino acid was
to check for the presence of an amino-acid substitution at the same position in the horn fly esterase, 11 clones resulting from the RT-PCR experiments on the diazinon-resistant Camp Cooley population sample were sequenced. No differences in nucleotide sequence of this 396 bp fragment were found between the susceptible and resistant populations. This fragment was used to probe a cDNA library from the diazinon-resistant Camp Cooley adult fly population, and a 2175 bp clone was isolated and designated HiaE7 (GenBank accession no.
AF139082). HiaE7 contains a 281 bp 59-untranslated
region, a 1713 bp open reading frame, a 181 bp 39 -untranslated region and a polyadenylated 39 end. The open reading frame is preceded by several in-frame stop codons and encodes a 570 amino-acid protein with 74% and 58% amino-acid identity to the LcaE7 and DmaE7
esterase-encoding genes from L. cuprina and D.
mel-anogaster, respectively. A polyadenylation signal sequence (AAUAAA) is located 140 bp downstream from the UAG stop codon. ASPCR assays of 50 individ-ual flies from the diazinon-resistant population showed no sequence differences at codon 137 of HiaE7 between
the susceptible and resistant flies (data not shown). The conceptual translation product of HiaE7 was
examined by alignment to the Torpedo californica ace-tylcholinesterase sequence (GenBank accession no. X03439) and found to contain 23 of the 24 invariant residues found in 29 lipases and esterases examined by Cygler et al. (1993), including three residues that could serve as the active-site motif, Ser218, G1u351 and His371
(data not shown). The contextual sequence surrounding these invariant residues was also conserved in HiαE7. The lone absent invariant residue was a cysteine puta-tively involved in disulfide bridge formation with Cys111.
However, there are two other cysteines at positions 88 and 89 in the primary sequence which could serve in bridge formation with Cys111. There are three regions of
HiαE7 that exhibit higher deviation from the amino-acid sequences of LcαE7 and DmαE7. These regions are Cys89
→Ser106, Met283→His295and Val358→Val437, which
have 56% and 44%, 62% and 38%, and 75% and 49% amino-acid identity plus conservative substitutions with LcαE7 and DmαE7, respectively. Outside these three regions HiαE7 exhibits 90% and 87% amino-acid ident-ity plus conservative substitutions with LcαE7 and DmαE7, respectively. Using the analysis of Cygler et al. (1993) and amino-acid alignment with the T. californica acetylcholinesterase, these three regions of HiαE7 corre-spond to regions noted as possessing high variability within the esterase family of enzymes. Cys89
→Ser106
corresponds to loop L1,2, a region near the conserved Trp
of the active site involved in substrate binding (Harel et al., 1993). Met283
→His295 appears to correspond to the
a3
6,7helix, while the long region between Val358→Val437
encompasses the a1–4
7,8 helices of which the a 1 7,8 and
a2
7,8helices display high variability among the esterases.
RT-PCR using FG-145 and FG-146 with poly(A)+ RNA from diazinon-resistant and diazinon-susceptible flies resulted in the amplification of a 326 bp cDNA (GenBank accession no. AF139081). This cDNA, desig-nated HiaE8, contained an open reading frame spanning
the entire 326 bp that encoded a protein with 84% and 75% amino-acid identity to the LcaE8 and DmaE8
esterase-encoding genes from L. cuprina and D.
mel-anogaster, respectively [Fig. 1(B)]. Eight independent
clones from the RT-PCR experiment with both the sus-ceptible and the resistant fly mRNA were sequenced and no nucleotide differences were found.
A 221 bp cDNA (GenBank accession no. AF139080) was amplified during the RT-PCR with 144 and FG-146 and RNA from diazinon-resistant flies. This cDNA, designated HiaE1, was found to have substantial DNA
sequence homology to a number of esterases from D.
melanogaster. The open reading frame from HiaE1
(data not shown) had from 55 to 60% amino-acid ident-ity to esterase-encoding genes DmaE1, DmaE2,
DmaE5, DmaE8, DmaE9 and DmaE10 from D.
mel-anogaster, and LcaE8 and LcaE9 from L. cuprina.
Although the highest amino-acid identity was to
DmaE1, no putative esterase designation could be
reliably ascertained from this short fragment. During the RNA Northern blot experiments, the HiaE1 cDNA
hybridized to a broad region of the Northern blot instead of a discrete band (data not shown). This result was later duplicated using a blot that had previously been shown to contain intact mRNA. Additionally, it was found that the HiaE1 probe could not be stripped from the blot
and interfered with subsequent reprobings with HiaE7.
Because of these difficulties, further investigations of the
HiaE1 cDNA were not done.
3.2. Analysis of HiaE7 and HiaE8 expression
To investigate possible resistance-associated quanti-tative differences in esterase gene expression, RNA from the susceptible and resistant populations was examined by Northern analysis. RNA was isolated from a sample of unfed, newly emerged, susceptible control flies, a second sample from the same population that had been fed for 3–5 days, and the diazinon-resistant Camp Cooley population. HiaE7 expression was elevated in
the resistant population compared with both susceptible samples (Fig. 2), while HiaE8 expression levels were
comparable in the unfed susceptible and the resistant Camp Cooley flies. HiaE8 expression was depressed in
the flies that had been fed for 3-5 days. HiaE7
hybridized to a single RNA species of approximately 2.8 kb in both the susceptible and the resistant samples, while the HiaE8 blot showed hybridization to two
Fig. 2. Northern analysis of RNA from diazinon-resistant and -sus-ceptible horn flies. Poly(A)+RNA was purified from susceptible (SUS) unfed, newly emerged adult horn flies (U), susceptible 3–5 day old flies fed citrated bovine blood (F), and diazinon-resistant (CC) adults of mixed age that had been feeding on cattle at the Camp Cooley ranch (F). The RNA was fractionated by denaturing formaldehyde gel electrophoresis and Northern blots probed with 32P-labeled 396 bp
HiaE7 (E7) and 326 bp HiaE8 (E8) RT-PCR cDNA fragments. 3µl and 1µg per lane of poly(A)+were used for the HiaE7 and HiaE8 blots, respectively. Autoradiography of both blots was under identical conditions using BioMax MS film and a Transcreen-HE intensifying screen (Kodak, Rochester, NY).
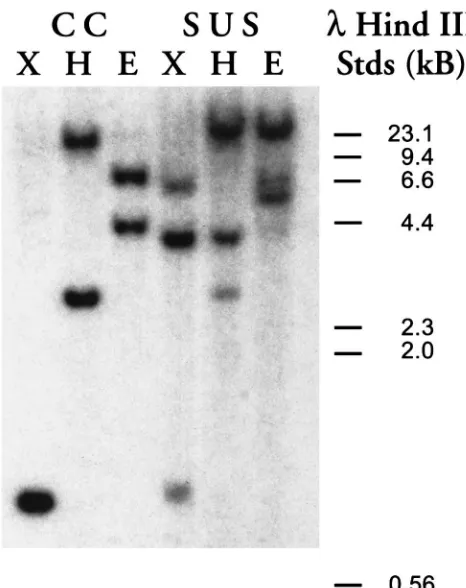
The enhanced expression of the HiaE7 transcript in
the resistant population could be due to an increase in gene copy number. To evaluate this possibility, restric-tion-enzyme-digested genomic DNA from both the sus-ceptible and resistant populations was fractionated by agarose gel electrophoresis and analyzed by Southern blotting. Probing the blot with the HiaE7 cDNA did not
reveal major differences between the OP-susceptible and OP-resistant samples, indicating a similar HiaE7 gene
copy number in both populations (Fig. 3).
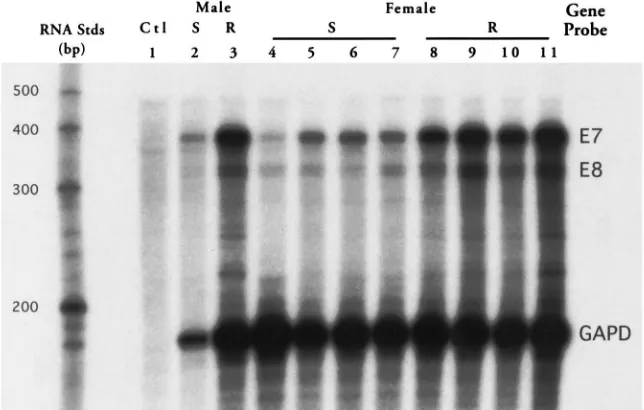
3.3. Ribonuclease protection assay of individual flies
Following the Northern analysis, which suggested an increase in HiaE7 expression in the resistant population,
coupled with information from previous studies which showed qualitative and quantitative variation in esterase protein expression among individual diazinon-resistant flies (Guerrero et al., 1999), individual fly HiaE7 gene
expression was assayed. Topical diazinon application to flies from the resistant population was used to phenotype individuals as diazinon-resistant or diazinon-susceptible. Total RNA was purified from one susceptible and resist-ant male, and four susceptible and resistresist-ant female flies, and probed in ribonuclease protection assays (Fig. 4). The susceptible male (lane 2) was one of four male indi-viduals that died approximately 2 h after the application of 5 ng of diazinon. Twenty-one male flies remained very active after 2 h at that dose. The resistant male (lane 3) survived 77 min after the application of 50 ng diazi-non. The other 24 flies in that treatment group died within 30 min after receiving this dose. The susceptible
Fig. 3. HiaE7 copy number analysis of horn fly genomic DNA. Gen-omic DNA (10µl) from diazinon-resistant (CC) and diazinon-suscep-tible (SUS) horn flies was digested with restriction enzyme XbaI (X), HindIII (H) or EcoRI (E), fractionated on a 0.7% agarose gel, trans-ferred to a nylon membrane by Southern blotting and probed with the
32P-labeled 396 bp HiaE7 RT-PCR cDNA fragment.
female flies in lanes 4 and 5 were two of the three indi-vidual females that were barely alive 2 h after receiving 5 ng of diazinon. The remaining 22 females that received this dosage were very active after 2 h. The susceptible females in lanes 6 and 7 died 17 and 24 min, respect-ively, after receiving 50 ng diazinon. Of the 55 female flies receiving the 50 ng dose, all but five died within 40 min. Resistant female flies in lanes 8, 9, 10 and 11 survived for 52, 47, 93 and 89 min, respectively, after receiving the 50 ng diazinon dose. The transcript levels of both HiaE7 and HiaE8 were elevated in the resistant
female individuals compared with the susceptible indi-viduals from the same population. The HiaE7 and
HiaE8 gene expression levels shown on the
autoradio-gram of Fig. 4 were quantified by densitometry and the values normalized by expressing the data relative to the expression of the GAPDH control hybridization probe (Table 2). There is about a five-fold increase in HiaE7
expression in the resistant females compared with the susceptible females, while HiaE8 is expressed about two
Fig. 4. RPA of HiaE7 and HiaE8 transcript levels in individual adult horn flies. Total RNA was purified from individual diazinon-resistant (R, lanes 3 and 8–11) and diazinon-susceptible (S, lanes 2 and 4–7) Camp Cooley male (lanes 2–3) and female (lanes 4–11) adult flies that were phenotyped by topical diazinon application. Each lane contained 0.5µg total RNA probed with 30,000 cpm each of32P-labeled antisense HiaE7
(E7), HiaE8 (E8) and GAPDH probes labeled to specific activities of 6.4×108cpm/µg, 5.3×108cpm/µg and 4.5×108cpm/µg, respectively.
Prelimi-nary experiments were performed to ensure the RPAs were done under conditions of probe excess. Lane 1 consists of a control reaction containing all three probes hybridized in the absence of RNA.
Table 2
Relative levels of HiaE7 and HiaE8 transcripts in individual diazinon-resistant (Res) and diazinon-susceptible (Sus) horn flies
Lane IDa Sex Phenotype E7b E8b
2 M Sus 0.41 0.11
3 M Res 0.43 0.009
4 F Sus 0.008 0.04
5 F Sus 0.09 0.06
6 F Sus 0.09 0.05
7 F Sus 0.08 0.06
Sus F mean (±standard deviation): 0.06 (±0.04) 0.05 (±0.01)
8 F Res 0.17 0.089
9 F Res 0.38 0.13
10 F Res 0.30 0.13
11 F Res 0.43 0.14
Res F mean (±standard deviation): 0.32 (±0.12) 0.12 (±0.02)
aFrom analysis of Fig. 4.
b Esterase mRNA transcript level relative to GAPDH.
4. Discussion
These investigations have resulted in the cloning of an esterase cDNA, HiaE7, that is the horn fly homolog
of the L. cuprina esterase-encoding gene, LcaE7. LcaE7
alleles from diazinon-resistant flies containing a Gly137
→Asp amino-acid substitution possess OP hydrolase
activity, leading to diazinon resistance in flies harboring the mutant allele (Newcomb et al., 1997a). HiaE7
expression was found to be higher in resistant horn flies and the elevated transcript levels did not appear due to an increased gene copy number in the resistant fly popu-lation. During the course of the sequencing, the portion
of the HiaE7 cDNA encompassing the region that would
contain the Gly137
→Asp substitution, if present, did not
appear to have sequence polymorphisms. However, it was possible that alleles of HiaE7 were present in the
resistant population at a low frequency and would only be detected by screening individual resistant flies for nucleotide polymorphisms in that region.
An ASPCR assay was developed to determine the sequence of codon 137 of the HiaE7 open reading frame
DNA template. Under these conditions primers FG-204 and FG-203 would successfully prime target DNA and yield amplification products only if the wild-type “sus-ceptible” nucleotide existed at the position of the codon 137’s first two and second nucleotides, respectively. The third nucleotide of the codon coding for Gly has four-fold redundancy and thus an assay for its identity was not developed. Fifty Camp Cooley flies were assayed and yielded positive ASPCR results with primer FG-204, indicating that these flies did not possess nucleotide sub-stitutions in the first two positions of codon 137. Ten of these 50 flies were further analyzed by ASPCR with pri-mers FG-209, FG-210 and FG-211, which differed from FG-204 only in the last nucleotide at the 39 end of the primer. These primers did not yield amplification pro-ducts, verifying the lack of sequence differences between individual flies at that position. These 10 flies were also assayed with FG-203, FG-206, FG-207 and FG-208. This set of four primers differed only in the 39-most nucleotide and would verify the identity of the second nucleotide of codon 137. Consistent with the results from the primer set described above, only FG-203, which contains the nucleotide coding for Gly at codon 137, yielded amplification.
From the information in this study, it does not appear that a mutated HiaE7 esterase predominates as a
diazi-non resistance mechanism. The five-fold elevated HiaE7
transcript levels of resistant female flies suggests that a sequestration mechanism may be important in the diazi-non resistance of these horn flies. Their higher level of HiαE7 compared with susceptible female flies will allow greater sequestration of insecticide, effectively pre-venting it from interacting with the target site. However, the ASPCR assay will not detect other mutations which might arise in HiaE7. Thus individuals from additional
OP-resistant populations must be identified and their
HiaE7 cDNAs sequenced to rule out a role for a mutated
HiaE7 in OP resistance. It is also likely that a
target-site insensitivity mechanism is contributing to diazinon resistance in horn flies. This mechanism could be mani-fested if the target of OP insecticides, acetylcholinester-ase, possessed amino-acid differences that affected the enzyme’s activity in the presence of OPs. Guerrero et al. (1999) reported that 40% of adult horn flies from Camp Cooley exhibited an OP-insensitive acetylcholine-sterase in dot blot assays. Cloning of acetylcholineacetylcholine-sterase from horn flies would be valuable to address the signifi-cance of this resistance mechanism.
There is some question about genetic differences between the susceptible laboratory colony flies and field fly populations, as the laboratory susceptible flies have been reared feeding on a cotton pad saturated with cit-rated bovine blood since 1961 without the introduction of new genetic material. Differences in gene expression between the laboratory colony and the Camp Cooley populations that do not seem related to diazinon
resist-ance have been noted (Guerrero and Kunz, 2000) and are possibly due to the loss of naturally occurring traits selected against by the laboratory rearing procedure. This was manifested in the HiaE8 polymorphism
between the susceptible laboratory colony flies and the Camp Cooley population (Fig. 2). The nature of the polymorphism is not known. However, a preliminary RPA experiment probing mRNA from male and female flies of various laboratory and field populations was con-ducted. Results indicated that the HiaE8 transcript from
the susceptible laboratory colony females was slightly larger than the transcript from the susceptible laboratory colony males (data not shown). The corresponding tran-script from the Camp Cooley flies was intermediate in size with no difference between males and females evi-dent. Although more definitive experiments need be done to verify this finding, a sex-related difference in transcript size would explain the HiaE8 polymorphism
of Fig. 2. It is also possible that heterogeneity in tran-scription start site or alternative splicing could be the cause of the polymorphism. It is known that the labora-tory susceptible population has lost the ability to enter diapause and that wild flies brought from field locations into the laboratory do not survive or reproduce well while feeding on citrated blood. These questions about the genetics of the laboratory-reared susceptible flies led to the topical diazinon selection experiments to allow a comparison of HiaE7 mRNA levels in resistant and
susceptible flies from the Camp Cooley population. In that manner, HiaE7 would exist in a similar genetic
background in the two groups of flies, differing primarily in their resistance to diazinon. In a set of preliminary experiments examining HiaE7 mRNA level in horn fly,
RPAs were performed on samples from eight different populations with varying degrees of diazinon sensitivity (data not shown). Each sample consisted of 25 flies of the same sex. The laboratory susceptible males did not show a detectable transcript which hybridized to the
HiaE7 probe, while the females of that population
showed only low levels of HiaE7 transcript relative to
other samples. It is not known if this is a trait associated with the OP susceptibility of that population or if it is due to loss or divergence of the allele from the years of laboratory rearing. All other samples had detectable hybridization to HiaE7, even male samples from
detection method to quantify levels of HiαE7 in individ-ual flies. Additional sampling of field populations with topical OP bioassays to identify OP-resistant individual flies and testing with the ELISA assay will help refine the relationship between OP resistance and HiαE7 in the horn fly.
Acknowledgements
I thank Mary Brumley for assistance with DNA clon-ing and sequencclon-ing, Kevin Temeyer for HiaE7 DNA
preparation and Matt Waldon for horn fly rearing and colony maintenance.
References
Ausubel, F.A., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K., 1998. In: Current Protocols in Molecular Biology. John Wiley & Sons, New York.
Burns, E.C., Wilson, B.H., 1963. Field resistance of horn flies to the organic phosphate insecticide ronnel. J. Econ. Entomol. 56, 718. Byford, R.L., Craig, M.E., DeRouen, S.M., Kimball, M.D., Morrison,
D.G., Wyatt, W.E., Foil, L.D., 1999. Influence of permethrin, diazi-non, and ivermectin treatments on insecticide resistance in the horn fly (Diptera: Muscidae). Int. J. Parasitol 29, 125–135.
Campbell, P.M., Newcomb, R.D., Russell, R.J., Oakeshott, J.G., 1998. Two different amino acid substitutions in the ali-esterase, E3, con-fer alternative types of organophosphorus insecticide resistance in the sheep blowfly, Lucilia cuprina. Insect Biochem. Mol. Biol. 28, 139–150.
Claudianos, C., Russell, R.J., Oakeshott, J.G., 1999. The same amino acid substitution in orthologous esterases confers organophosphate resistance on the house fly and a blowfly. Insect Biochem. Mol. Biol. 29, 675–686.
Conyers, C.M., MacNicoll, A.D., Price, N.R., 1998. Purification and characterization of an esterase involved in resistance to organ-ophosphorus insecticides in the saw-toothed grain beetle,
Oryzae-philus surinamensis (Coleoptera: Silvanidae). Insect Biochem. Mol. Biol. 28, 435–448.
Cygler, M., Schrag, J.D., Sussman, J.L., Harel, M., Silman, I., Gentry, M.K., Doctor, B.P., 1993. Relationship between sequence conser-vation and three-dimensional structure in a large family of ester-ases, lipester-ases, and related proteins. Protein Sci. 2, 366–382. Guerrero, F.D., Kunz, S.E., 2000. Laboratory rearing conditions select
for differences in gene expression between laboratory and wild type horn flies. Southwest. Entomol., in press.
Guerrero, F.D., Pruett, J.H., Kunz, S.E., Kammlah, D.M., 1999. Ester-ase profiles of diazinon-susceptible and -resistant horn flies (Diptera: Muscidae). J. Econ. Entomol. 92, 286–292.
Harel, M., Schalk, I., Ehret-Sabatier, L., Bouet, F., Goeldner, M., Hirth, C., Axelsen, P.H., Silman, I., Sussman, J.L., 1993. Quatern-ary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc. Natl. Acad. Sci. USA 90, 9031–9035. Hecker, K.H., Roux, K.H., 1996. High and low annealing temperatures
increase both specificity and yield in touchdown and stepdown PCR. Biotechniques 20, 478–485.
Hemingway, J., Karunaratne, S.H.P.P., 1998. Mosquito carboxylester-ases: a review of the molecular biology and biochemistry of a major insecticide resistance mechanism. Med. Vet. Entomol. 12, 1–12. Newcomb, R.D., Campbell, P.M., Ollis, D.L., Cheah, E., Russell, R.J.,
Oakeshott, J.G., 1997a. A single amino acid substitution converts a carboxylesterase to an organophosphorus hydrolase and confers insecticide resistance on a blowfly. Proc. Natl. Acad. Sci. USA 94, 7464–7468.
Newcomb, R.D., Campbell, P.M., Russell, R.J., Oakeshott, J.G., 1997b. cDNA cloning, baculovirus-expression and kinetic proper-ties of the esterase, E3, involved in organophosphorus resistance in Lucilia cuprina. Insect Biochem. Mol. Biol. 27, 15–25. Scharf, M.E., Hemingway, J., Small, G.J., Bennett, G.W., 1997.
Exam-ination of esterases from insecticide resistant and susceptible strains of the german cockroach, Blattella germanica (L.). Insect Biochem. Mol. Biol. 27, 489–497.
Schmidt, C.D., Dreiss, J.M., Eschle, J.L., Harris, R.L., Pickens, M.O., 1976. Horn fly: modified laboratory rearing methods. Southwest. Entomol. 1, 49–51.
Sparks, T.C., Quisenberry, S.S., Lockwood, J.A., Byford, R.L., Roush, R.T., 1985. Insecticide resistance in the horn fly, Haematobia irrit-ans. J. Agric. Entomol. 2, 217–233.