Recently, it has been shown that the same sets of genes act in both root and shoot to regulate cell fate and patterning. One gene cassette regulates epidermal cell fate, another cassette regulates ground tissue derived cell fate and organization. Ectopic expression and laser ablation have been used to probe the mechanisms by which these genes perform their tissue and organ-specific functions.
Addresses
Biology Department, New York University, 1009 Main Building, New York, NY 10003, USA; e-mail: [email protected]
Current Opinion in Plant Biology1999, 2:39–43 http://biomednet.com/elecref/1369526600200039 © Elsevier Science Ltd ISSN 1369-5266
Abbreviations H hair cells
N non-hair cells
Introduction
Viewed in gross outline, the root and shoot could be mir-ror images: both are ramified structures — one exploring the ground, the other the air. But at closer inspection their differences seem to far outweigh their similarities. Roots are among the simplest organs in their morphology, shoots produce highly complex organs; roots branch from internal differentiated tissues, shoots set aside axillary meristems at each leaf; the root meristem is covered by the root cap, the shoot meristem is exposed on its apical surface. Initial genetic data have also emphasized the differences between roots and shoots — mutations that have dramat-ic effects on the shoot meristem, have no apparent effect on the root. These are only a few of the differences that would suggest that shoots and roots are unlikely to share common mechanisms regulating their patterning, growth or differentiation.
Several recent findings in Arabidopsis have challenged this view. Two independent sets of genes have now been shown to control aspects of shoot and root cell fate and patterning. One regulates the pattern and differentiation of epidermal cells, the other the radial pattern of ground tissue. Here, I will review current models of how these genes interact with each other, then address attempts to uncover the factors responsible for tissue-specific fate determination.
Epidermal patterning in root and shoot
The root epidermis is composed of two cell types, hair cells (H) and non-hair cells (N). Three genes have been identified by mutation as controlling the fate of individual epidermal cells. GLABRA 2 (GL2) is a negative regulator of H fate; where it is active, cells take on an N fate. GL2encodes a homeobox-containing putative transcription factor and is
expressed preferentially in non-hair cells [1–3]. TRANSPAR-ENT TESTA GLABRA(TTG) acts as an activator of GL2and thus also is a negative regulator of H fate [2,4,5•]. Mutation of either ttgor gl2results in ectopic root hairs. Cloning of TTG has not yet been reported. Mutations in CAPRICE (CPC) have the opposite phenotype to ttg and gl2— H cell produc-tion is dramatically reduced. Genetic and molecular data indicate that CPCis a negative regulator of GL2, making it formally a positive regulator of H fate [6••]. CPCencodes a myb-family protein. The current model then is that CPC and TTG act independently on GL2: CPC represses, whereas TTG activates it (Figure 1).
The shoot epidermis is more complex than that of the root as it contains two specialized cell types, stomata and trichomes. Both gl2 and ttg were first identified by the absence of normal trichomes in the leaf [7,8]. Trichomes are similar to root hairs in that projections are formed from a single epidermal cell. The root phenotype of excess root-hairs, however, appears to be opposite to the shoot phenotype on the same mutant of loss of trichomes. Nevertheless, the similarities in the pathways are strik-ing. For trichome formation, TTGis a positive regulator of GL2[2,5•,9]. Upstream of GL2there is also a myb- fami-ly gene, GLABRA 1 (GL1) [10,11] (Figure 1). Although evidence for CPC expression in the shoot has not yet been found, overexpression of CPC results in a strong reduction in trichome number [6••]. This could be due to it interacting with a natural target, or its myb-domain may be sequestering GL2 in the shoot, resulting in reduced trichome formation.
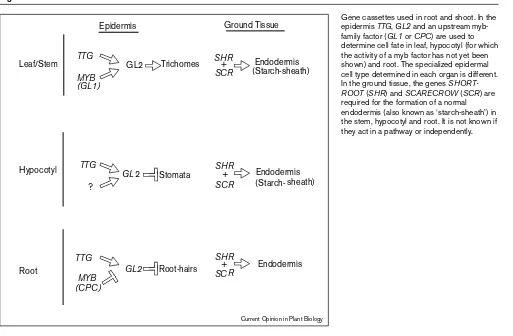
Examination of the expression pattern of GL2 in the hypocotyl led to the discovery that the patterning of stom-ata in the hypocotyl is also dependent on TTG and GL2[5•] (Figure 1). It was found that stomata are located in the same relative position in relation to the underlying cortex cells as are root hairs and mutations in either gl2 or ttg resulted in ectopic localization of stomata. Moreover, a 500 bp promoter region was shown to be essential for the cell-specific expression pattern of GL2 in both root and hypocotyl [5•]. This region contains a putative myb-bind-ing site and the same region is important for trichome expression of GL2[12•]. In summary, both shoot and root share a pathway for epidermal cell development that includes the genes TTGand GL2as well as an upstream myb-family factor.
Ground tissue patterning in root and shoot
A second example of a set of genes that appears to perform the same function in root and shoot comes from the analy-sis of ground tissue formation. In the root, the ground tissue is separated into an outer cortex and an inner endo-dermis [13]. In bothshort-root(shr) and scarecrow(scr), thereIs the shoot a root with a view?
is a only a single ground tissue-derived layer in the root [14,15]. Analysis of tissue-specific markers showed thatshr is missing endodermal attributes in the remaining layer, whereas in scrthis layer has attributes of both cortex and endodermis [14,16]. This suggests that SHRis required for endodermal fate specification, whereas SCRis involved in effecting the asymmetric division that the initial cell undergoes to form the two cell layers (Figure 1). The SCR gene encodes a protein with motifs suggestive of a tran-scription factor [16].
The shoot phenotype of both shrand scrmutants is a loss of the normal gravitropic response in the hypocotyl and inflorescence [17••]. The apparent cause is the absence of the normal endodermis, also known as the starch sheath which has been hypothesized to be the site of gravitropic sensing in the shoot (Figure 1).
The hypocotyl endodermis is formed during embryogene-sis concurrently with the root endodermis through a series of divisions of the ground tissue. Embryonic analysis has shown that SCR and SHR function are required for this division process [15] (Y Helariutta and PN Benfey, unpub-lished data). Formation of the endodermis from the root meristem, however, appears to be quite different from the process that occurs in the shoot meristem. Fate mapping
has shown that the formation of endodermis and cortex in the root requires two asymmetric divisions of the cortex/endodermal initial cell. This cell is found at the root tip where the cortex and endodermal cell files converge. Similarly to animal stem cells, it first divides to regenerate an initial and then the upper daughter divides to form the progenitors of the cortex and endodermal cell files [13]. Fate mapping of the internal shoot tissues including the endodermis has not yet been performed. Anatomical analysis indicates that the shoot endodermis originates as a layer of cells surrounding the vascular strands that traverse the petiole. When these vascular strands join the stem the endodermis forms a single layer surrounding all of the vas-cular bundles. Future analysis of SCRand SHRactivity in the shoot, combined with a better understanding of the ontogeny of shoot ground tissue should determine how similar the actual mechanism is by which the ground tissue is patterned in both shoot and root.
Determining that a set of genes performs similar devel-opmental tasks in more than one context (tissue, organ etc.) is viewed as evidence for a common evolutionary origin [18]. It has been hypothesized that present-day shoots and roots evolved from a common ‘ancestral meristem’ [19]. Alternatively, tissue-specific patterning and cell fate specification may have predated the first
Figure 1
Gene cassettes used in root and shoot. In the epidermis TTG, GL2and an upstream myb-family factor (GL1or CPC) are used to determine cell fate in leaf, hypocotyl (for which the activity of a myb factor has not yet been shown) and root. The specialized epidermal cell type determined in each organ is different. In the ground tissue, the genes SHORT-ROOT(SHR) and SCARECROW (SCR) are required for the formation of a normal endodermis (also known as ‘starch-sheath’) in the stem, hypocotyl and root. It is not known if they act in a pathway or independently.
Hypocotyl
Root
Ground Tissue Epidermis
(GL1) TTG
GL2
? TTG
GL2
GL2
Stomata Trichomes
TTG
(CPC)
Root-hairs
Leaf/Stem SHR+ Endodermis
SCR (Starch-sheath)
Endodermis SHR
+ SCR
Endodermis SHR
+ SCR
(Starch- sheath) MYB
MYB
meristem and thus may have been incorporated inde-pendently as the root and shoot meristems evolved. What is not clear is how many mechanisms are shared. Further evidence in support of at least some common mechanisms is the isolation of the Defective embryo and meristems (Dem) gene which is required for both shoot and root meristem function in tomato and is expressed in both meristems [20]. Nevertheless, mutational analysis has also provided evidence for specialized functions that are found in one but not the other meristem. To date no root phenotype has been found to be associated with the mutation of the shoot meristemlessgene [21] which, as its name suggests, results in loss of a functioning shoot meristem, nor has one been found with the wuschel[22] or zwille [23] mutants in which the shoot meristem is severely compromised. It is possible that there are homologs that perform similar functions in the root, or that these genes are specialized for the shoot meristem’s job of creating lateral appendages (leaves) for which there is no equivalent in the root.
Organ and tissue-specific cell fate
The idea has been presented that the shoot and root use the same gene cassettes to perform similar functions. Although the MYB/TTG/GL2 cassette has been shown to regulate the placement and fate of specialized epidermal cells in the root, hypocotyl and leaf, the resulting cell type is different for each of these organs. Moreover, these genes negatively regulate the specialized epidermal cells in the root and hypocotyl, and positively regulate tri-chome fate in the leaf. What can account for the differences in activity of the same genes in different organs? Part of the answer lies in the fact that some of the genes in the cassette are transcription factors which could interact with different downstream targets, activating the genes required for trichome initiation in leaves and repressing the genes for root hair or stomatal initiation in root and hypocotyl.
One approach to identifying which of the genes in the pathway controls organ- and tissue-specific cell fate determination is to ectopically express each of the genes and ask if this is sufficient to produce the cell type in an abnormal location. As mentioned above, CPC ectopic expression results in severely reduced trichome numbers and ectopic root hairs [6••]. Because there is no change in specialized cell type in either shoot or root (i.e. root hairs aren’t produced on leaves), nor does ectopic expression cause a change in the tissue or organ-specificity of tri-chome formation in the shoot, it is unlikely that CPC plays a role in determining organ- or tissue-specificity. It has been shown that the maize myb-domain containing R gene, which is involved in anthocyanin biosynthesis, can rescue the ttg mutant phenotype indicating that it can substitute for the Arabidopsis TTG gene. Overexpression of Rresults in reduced root hair numbers and trichome formation on shoot organs on which trichomes are not normally found [24].
This suggests that TTG regulates the organ specificity of trichome formation in the shoot, but that it does not control the type of specialized cell made in the shoot versus in the root. Initial results indicated that ectopic expression of GL1 causes a reduction in trichome number [10], which is con-trary to what would be expected of an activator of GL2. Recently, ectopic expression of GL1 has been analyzed in plants in which the tryptichon gene (a negative regulator of trichome formation) was mutated. This resulted in addi-tional leaf-borne trichomes as well as trichome formation in the sub-epidermal layers of the leaf with similar patterning to that found in a normal leaf epidermis [25••]. This indi-cates that GL1 regulates the tissue-specific formation of trichomes. Unexpectedly, these plants also had trichomes on several stem derived organs (pistils, stamens, flower stalks) which normally do not have trichomes [25••]. Because previous work pointed to TTGas the prime regu-lator of shoot organ-specificity, it has been suggested that overexpression of GL1might induce TTG in these organs [25••]. Nevertheless, in both of the GL1overexpression sit-uations (with or without mutant tryptichon), no alteration in root hair number or placement was observed [10,25••].
These results present the following picture of the activities of each of these genes: GL1 is normally expressed only in the epidermis and is present in many shoot organs at low levels but not in roots [10,11,12•,25••]; TTG is expressed only in specific shoot organs (e.g. leaves and hypocotyls) and is limiting for trichome formation. Overexpression of GL1can induce TTGexpression in shoot organs in which it is not normally expressed. Because overexpression of GL1 has no effect on root epidermal cell fate, and overexpres-sion of a TTGsubstitute or CPConly alters the amount but not the type of cell (root-hair versus trichome), other, as yet unidentified, factors must be present to distinguish the root from the shoot pathways. These results indicate that the decision to make a specialized epidermal cell in the root and shoot involves multiple steps. As the molecules are identified that regulate this pathway, it should become clear to what extent these decisions are made in a hierar-chical manner (for example: epidermis, then shoot versus root, then which shoot organ) as opposed to overlapping inputs that are integrated by the downstream targets of each of these genes.
Signaling between cells
laser ablation was used to isolate epidermal cells from their epidermal neighbors or from the underlying cortex cells. The surprising finding was that there was no change in cell fate with either type of ablation [26••]. As it was not technically feasible to completely isolate an epidermal cell from both cortex and epidermal neighbors, it is possible that redundant pathways exist. An alternative possibility is that positional information is not maintained by cell–cell communication at all, but rather comes from factors found in the cell walls underneath the cells. Support for this hypothesis comes from analysis of the first division of the brown alga, Fucus, in which cell wall factors were shown to determine cell fate [27]. A recent analysis of later stages of Fucus development, however, indicates that cell wall factors do not play a part in fate determination [28], suggesting that this is not a general mechanism in plants.
Conclusions
The finding that gene cassettes are providing a similar developmental function in shoot and root should not come as a surprise to plant evolutionary biologists for whom it has always appeared likely that the root meristem evolved from an ancestral shoot. Developmental geneticists, on the other hand, have generally separated the plant into root and shoot processes and until recently these have appeared to be quite different. The notable exceptions have been that in shoot and root, cell fate is determined by positional information and that the core function of both meristems is to maintain an undifferentiated popula-tion of cells from which new tissues can be formed. Future work should include determining how many other con-served gene cassettes act in both shoot and root. A central focus should be on defining the mechanisms by which these genes act and are acted upon to determine pattern-ing and cell fate in different developmental contexts. These are exciting times, as we are rapidly closing in on the molecular mechanisms that regulate the most funda-mental processes of plant development.
Acknowledgements
I would like to thank Ben Scheres and Jocelyn Malamy for very helpful discussions and comments. Support for work in my laboratory on radial pattern formation is provided by a grant from the NIH(GM#R01-43772) and from the Human Frontiers Science Program.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest ••of outstanding interest
1. Rerie WG, Feldmann KA, Marks MD: The GLABRA2 gene encodes a homeo domain protein required for normal trichome
development in Arabidopsis.Genes Dev1994, 8:1388-1399. 2. Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I,
Morelli G: The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development.Plant J 1996, 10:393-402.
3. Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW: The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of
Arabidopsis thaliana.Development1996, 122:1253-1260.
4. Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein: The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root.Dev Biol1994, 166:740-754. 5. Hung CY, Lin Y, Zhang M, Pollock S, Marks MD, Schiefelbein J:
• A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis.Plant Physiol1998, 117:73-84.
This paper is of interest because it provides the first evidence that stom-atal patterning in the hypocotyl is under the same control of the TTG/GL2 cassette as root hair patterning.
6. Wada T, Tachibana T, Shimura Y, Okada K: Epidermal cell
•• differentiation in Arabidopsis determined by a Myb homolog, CPC. Science1997, 277:1113-1116.
This paper presents a major breakthrough in our understanding of root hair formation through the cloning and characterization of a new myb-family gene, CPC which is shown to be an upstream regulator of root-hair formation and GL2 expression.
7. Koornneef M, Dellaert SWM, van der Veen JH: EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L) Heynh.Mutat Res 1982, 93:109-123.
8. Koornneef M: The complex syndrome of ttg mutants.Arabidopsis Inf Serv1981, 18: 45-51.
9. Hulskamp M, Misra S, Jürgens G: Genetic dissection of trichome cell development in Arabidopsis.Cell1994, 76:555-566. 10. Larkin JC, Oppenheimer DG, Lloyd AM, Paparozzi ET, Marks MD:
Roles of the glabrous1and transparent testa glabragenes in
Arabidopsis development.Plant Cell1994, 6:1065-1076. 11. Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD: A
myb gene required for leaf trichome differentiation in Arabidopsis
is expressed in stipules.Cell1991, 67:483-493.
12. Szymanski DB, Jilk RA, Pollock SM, Marks MD: Control of GL2
• expression in Arabidopsis leaves and trichomes.Development 1998, 125:1161-1171.
Further evidence that shoot and root use the same pathway comes from the finding that the same promoter region responsible for root and hypocotyl tis-sue-specific expression is required for trichome-specific expression.
13. Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B:Cellular organization of the Arabidopsis thaliana root. Development1993, 119:71-84.
14. Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser M-T, Aeschbacher RA: Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis.Development1993, 119:57-70.
15. Scheres B, Di Laurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey PN:Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the radial axis.Development1995, 121:53-62.
16. Di Laurenzio L, Wysocka-Diller J , Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN: The SCARECROW
gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root.Cell 1996, 86:423-433.
17. Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M:
•• Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana.Plant J1998, 14:425-430. Addressing the long-standing question of the site of gravitropic sensing in the shoot led to the discovery that mutations in the root radial pattern regu-lators,SCARECROWand SHORT-ROOTalso cause the loss of a normal endodermis in the hypocotyl and shoot inflorescence.
18. Tautz D: Evolutionary biology. Debatable homologies.Nature1998, 395:17-19.
19. Steeves TA, Sussex IM:Patterns in Plant Development.Cambridge: Cambridge University Press, 1991.
20. Keddie JS, Carroll BJ, Thomas CM, Reyes ME, Klimyuk V, Holtan H, Gruissem W, Jones JDG: Transposon tagging of the Defective embryo and meristems gene of tomato.Plant Cell1998, 10:877-888.
21. Long JA, Moan EI, Medford JI, Barton MK: A member of the KNOTTED class of homeodomain proteins encoded by the STM
gene of Arabidopsis.Nature1996, 379:66-69.
23. Moussian B, Schoof H, Haecker A, Jurgens G, Laux T: Role of the
ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis.EMBO J1998, 17:1799-1809. 24. Lloyd AM, Walbot V, Davis RW: Arabidopsis and Nicotiana
anthocyanin production activated by maize regulators R and C1. Science1992, 258:1773-1775.
25. Schnittger A, Jurgens G, Hulskamp M: Tissue layer and organ •• specificity of trichome formation are regulated by GLABRA1 and
TRIPTYCHON in Arabidopsis.Development1998, 125:2283-2289. This paper reports the surprising finding that when ectopically expressed in a tryptichon mutant background, GL1 is able to cause the formation of tri-chomes on various shoot organs and in the subepidermal tissue of leaves. Nevertheless, no alteration of root epidermal cell fate is observed.
26. Berger F, Haseloff J, Schiefelbein J, Dolan L: Positional information •• in root epidermis is defined during embryogenesis and acts in
domains with strict boundaries.Curr Biol1998, 8:421-430. Counter to expectation, this paper reports that laser ablation of the cells sur-rounding root epidermal cells results in no detectable change in cell fate, leading the authors to postulate that factors in the underlying cell walls are responsible for fate determination.
27. Berger F, Taylor AR, Brownlee C: Cell fate determination by the cell wall in early Fucus development.Science1994, 263:1421-1423.