Answer Key
Unit 3 Molecular Genetics
Answers to Unit 3 Preparation Questions
Assessing Student Readiness
(Student textbook pages 198–201)
1.
Characteristic Prokaryotes Eukaryotes
Relative cell size small large
Cell number in typical organism
single multiple
Location of genetic material cytoplasm nucleus
Membrane-bond genetic material
no yes
Number of chromosomes one several
2. c
3. Any three of: they are not cells; they have no
cytoplasm, cell membrane, or organelles; they cannot reproduce outside of host cells; they are dormant outside of host cells.
4. c
5. d
6. d
7. a. endoplasmic reticulum (ER)
b. Synthesis of proteins and synthesis of lipids and lipid-containing molecules. For example, in the liver, the ER helps detoxify the blood of drugs and alcohol. In the testes and ovaries, the ER produces testosterone and estrogen.
8. c
9. e
10. d
11. Enzymes help facilitate chemical reactions by acting as protein catalysts that increase the rate of the reaction. 12. hese indentations (active sites) facilitate
substrate-enzyme binding as the active site changes in shape to accommodate the substrate in what is called induced it.
13. Enzymes are classiied according to the type of reactions they catalyze.
14. nucleotide → nucleic acid → gene → protein Nucleotides are the building blocks of nucleic acids, which make up genes, which code for proteins. 15. d
16. a. Hydrogen bonds link nucleotides together between the two strands that make up a DNA molecule. hese bonds occur between complementary bases. b. he covalent bonds that link adjacent nucleotides
together within each strand are called
phosphodiester bonds. hese bonds occur between the phosphate group on one nucleotide and a hydroxyl group on the sugar of the next nucleotide in the same strand.
17. e
18. An allele is a diferent form of the same gene. Homologous chromosomes carry two alleles of the same gene. Diferences in these alleles account for diferences in hair colour.
19. a
20. a
21. e
22. • Deletion—a piece of a chromosome is cut out
• Duplication—a piece of a chromosome appears twice or more
• Inversion—a piece of a chromosome is lipped • Translocation—a segment of a chromosome is
attached to another chromosome 23. a. prophase I
b. crossing over
c. During crossing over, the chemical bonds that hold the DNA together in the chromosome are broken and reformed. In some cases, the chromosomes do not reform correctly.
24. he chromosome errors described result in genetic disorders when they delete, alter, or duplicate genes. In some case, however, the errors afect regions of the genome that lack genes or are within non-coding regions of a gene. hese errors are not harmful.
26. Sample answer: Genetic engineering can create transgenic organisms that secrete a human protein for medical use. First, the human gene that codes for the protein is injected into an egg from a donor goat. hen the egg is placed in a host goat where a transgenic goat develops. Finally, this transgenic goat produces milk containing the human hormone.
27. c
28. Viruses enter host cells and direct the activity of their host cell’s DNA. his makes viruses useful tools for producing a copy of a gene. Researchers can insert the gene of interest into the virus genome. he virus then directs the host cell to make multiple copies of the virus; each new virus that the cell produces will contain the gene of interest as well.
29. a. transgenic or genetically modiied organism b. It has a diferent genotype, as it now contains four
new genes
c. It has a diferent phenotype, as it now displays higher levels of iron, vitamin A, sulfur, and a new enzyme.
Chapter 5 The Structure
and Function of DNA
Answers to Learning Check Questions
(Student textbook page 207)1. Genetic material must contain information that regulates the production of proteins. It also must be able to accurately replicate itself to maintain continuity in future generations. Genetic material must allow for some mutations so that there is variation within a species.
2. Griith used two forms of S. pneumoniae: a pathogenic S-strain and a non-pathogenic R-strain. Ater injecting mice with a mixture of heat-killed S-strain and live R-strain, the mice died. Griith concluded that something from the heat-killed S-strain transferred to the R-strain to transform it into a pathogenic form.
3. • When they treated heat-killed pathogenic bacteria with a protein-destroying enzyme, transformation still occurred.
• When they treated heat-killed pathogenic bacteria with a DNA-destroying enzyme, transformation did not occur. hese results provided strong evidence for DNA’s role in transformation.
4. Two diferent radioactive isotopes were used to trace each type of molecule. One sample of T2 virus was tagged with radioactive phosphorus (32P), since phosphorus is present in DNA and not protein. he other sample of T2 virus was tagged with radioactive sulfur (35S), since sulfur is
only found in the protein coat of the capsid.
5. he independent variable in the experiment was the type of radioactive isotope used to tag the virus. he dependent variable in the experiment was the presence of radioactivity inside the infected bacterial cells. Controls include the usage of the same type of virus in both experiments and the usage of the same protocol for infecting bacterial cells in both experiments.
6. Bacterial cells that are infected by viruses with
32P-labelled DNA would not be radioactive. Bacterial
cells infected by viruses with 35S-labelled capsid proteins would be radioactive.
(Student textbook page 212)
7. Answers should resemble Figure 5.4 on page 208 of the student textbook, with labels for phosphate group, sugar group, and nitrogen-containing base.
8. Nucleotides in DNA have a deoxyribose sugar, while nucleotides in RNA have a ribose sugar with a hydroxyl group at carbon 2. In addition to the sugar group, each nucleotide is attached to a phosphate group and a base. he bases are adenine, cytosine, guanine, and thymine in the case of DNA, and adenine, cytosine, guanine, and uracil in the case of RNA.
9. Chargaf ’s rule states that, in the DNA nucleotides, the amount of adenine will be more or less equal to the amount of thymine, and the amount of guanine will be equal to the amount of cytosine. he number of A-T nucleotides will not necessarily equal the number of C-G nucleotides. his overturned Levene’s earlier hypothesis that the nucleotides occurred in equal amounts and were present in a constant and repeated sequence.
11. Diagrams should resemble Figure 5.7B on page 213 of the student textbook. Base pairing and directionality of strands should be shown.
12. Nucleic acids are soluble in water. herefore, the nitrogenous bases, which are somewhat hydrophobic, must be positioned away from the water found in the nucleoplasm, and the polar phosphate groups (which are hydrophilic) must be on the outside of the molecule, interacting with the water.
(Student textbook page 222)
13. he main objective of DNA replication is to produce two identical DNA molecules from a parent DNA molecule.
14. DNA replication occurs during the S phase of interphase, prior to cell division, ensuring that there is a copy available for each new daughter cell. 15. • Conservative model—Two new daughter strands
form to create a new double helix, and the original DNA strands re-form into the parent molecule. • Semi-conservative model—Each new DNA molecule
contains one strand of the original DNA and one newly synthesized strand.
• Dispersive model—Parental DNA is broken into fragments. herefore, the daughter DNA contains a mix of parental and newly synthesized DNA.
16. Nitrogen is a component of DNA and is incorporated into newly synthesized daughter strands. Having a “light” form (14N) and a “heavy” form (15N) allowed the separation of diferent DNA strands based on the amount of isotope present in the newly synthesized DNA. DNA with more 15N would be denser than DNA with 14N, and could therefore be separated by centrifugation.
17. Meselson and Stahl concluded that DNA replication is semi-conservative. Ater one round of replication, DNA appeared as a single band, midway between the expected positions of 15N-labelled DNA and 14N-labelled DNA. Ater the second round of
replication, DNA appeared as two bands, with one band corresponding to 14N-labelled DNA and the other band in the position of hybrid DNA (half 14N
and half 15N). In additional rounds of replication, the
same two bands were observed, therefore supporting the semi-conservative model.
18. Each new cell that is produced must have an exact copy of parental DNA. he daughter strands of DNA are part of a DNA molecule that will be in the daughter cells. his ensures that newly born cells are similar to parents and maintain their genetic identity.
(Student textbook page 227)
19. Initiation—Helicase enzymes unwind DNA to separate it into two strands. A replication bubble is formed when single-strand binding proteins stabilize the separated strands.
Elongation—New DNA strands are synthesized by joining free nucleotides together. his is catalyzed by DNA polymerase, which synthesizes the new strands that are complementary to the parental strand. Termination—he two new DNA molecules separate from one another.
20. Replication takes place in a slightly diferent way on each DNA strand because DNA polymerase can only catalyze elongation in the 5′ to 3′ direction. In order for both strands of DNA to be synthesized simultaneously, the method of replication must difer.
21. On the leading strand, DNA synthesis takes place along the DNA molecule in the same direction as the movement of the replication fork. On the lagging strand, DNA synthesis proceeds in the opposite direction to the movement of the replication fork. he lagging strand is synthesized in short fragments called Okazaki fragments.
22. DNA replication requires the use of many enzymes that have speciic roles. he presence of numerous specialized enzymes may relect the importance of having accurate DNA replication, since mutations in DNA can change the genetic makeup of an organism.
23. Answers may include: DNA polymerases have a proofreading function during which they excise incorrect bases and add the correct bases. Mismatch repair involves a group of enzymes that identify, remove, and replace incorrect bases.
24. Many tissues and organs require continuous cell regeneration. herefore, DNA replication must be quick and accurate so that new daughter cells receive exact copies of DNA from the parent cell.
Answers to Caption Questions
Figure 5.2 (Student textbook page 205): If a live strain had been transferred, the efects would have been due to that strain, not due to the transfer of a substance form it to the R-strain, which makes it pathogenic.
Figure 5.10 (Student textbook page 215): Twisting a rubber band around itself mimics how DNA supercoiling. he rubber band becomes compacted due to the coils that twisting forms. his model is also useful since it demonstrates the tension that is created by supercoiling. A rubber band may become linearized, where supercoiling in bacterial DNA occurs because it is a circular. he rubber band model also does not relect the double-stranded nature of DNA.
Figure 5.16 (Student textbook page 221): If DNA had not been uniformly labelled with 15N, the banding patterns
would not accurately relect the presence of parental DNA.
Answers to Section 5.1 Review Questions
(Student textbook page 218)1. Griith’s experiments showed the existence of a transforming principle. hat is, something in the heat-killed pathogenic bacteria (S-strain) could transform the non-pathogenic bacteria (R-strain) into a pathogenic form. his result led to Avery’s experiments on
Streptococcus pneumoniae to identify the molecules that caused this transformation. Avery’s research concluded that DNA was the transforming principle.
2. a. Graphic organizers should include information about experimental setup (i.e., the use of two radioactive isotopes to diferentially label DNA and capsid protein), experimental procedure (i.e., the use of agitation in a blender to dislodge viruses, and subsequent centrifugation), and results. A summary of the experiment is found in Figure 5.3 on page 207 of the student textbook.
b. he results showed that DNA is the hereditary material.
3. Miescher isolated nuclein from the nucleus of white blood cells. He found that this material was present only in the nuclei of cells. Further experimentation showed that nuclein was a weakly acidic phosphorus-containing substance. Nuclein would later be known as nucleic acid or, more speciically, DNA (deoxyribonucleic acid).
4. Diagrams should resemble the marginal portion of Figure 5.4 on page 208 of the student textbook.
5. he nucleotide composition of the human would be diferent from the nucleotide composition of the mouse because the composition of DNA is unique to each species. However, the percentage of adenine will remain approximately the same as the percentage of thymine, and the percentage of cytosine will remain approximately equal to the percentage of guanine in each species.
6. C = 26%; G = 26%; T = 24%
7. Diagrams should include labels for sugar-phosphate molecules (“handrails”), nucleotide base pairing (“rungs”), and directionality of both strands and resemble the close up of Figure 5.7 on page 213 of the student textbook.
8. a. Levene proposed that DNA was composed of nucleotides, and that each of the four types of nucleotides contained one of four nitrogen-containing bases, a sugar molecule, and a phosphate group. b. Chargaf showed that DNA is composed of repeating
units of nucleotides in ixed proportions (i.e., the percent composition is of adenine is the same as thymine, and the percent composition of cytosine is the same as guanine). Chargaf ’s rule helped Watson and Crick infer that adenine paired with thymine, and cytosine paired with guanine.
c. Franklin determined that DNA had a helical structure, with nitrogenous bases located on the inside of the structure, and the sugar-phosphate backbone located on the outside. his information led to Watson and Crick’s ladder-like double helix model of DNA, with the sugar-phosphate molecules acting as “handrails” and the bases making up the “rungs.” d. Pauling discovered that proteins have a helical
structure. his discovery inluenced Watson and Crick to propose that DNA was shaped like a helix. 9. a. 5′-ATTGAACAT-3′
b. 5′-GATTAACGG-3′
c. 5′-CGGAGCTAA-3′
10. A gene is a functional unit of DNA. It is a speciic sequence that encodes for proteins or RNA molecules. A genome is an organism’s complete genetic makeup. It is composed of an organism’s total DNA sequence. 11. Venn diagrams should include:
• Prokaryotes Only—double-stranded, circular DNA packed in the nucleoid; DNA is compacted via supercoiling; most are haploid; genomes contain very little non-essential DNA; contain plasmids
• Prokaryotes and Eukaryotes—chromosomal DNA is much larger than their cells, and therefore must be compacted
12. Diagrams should include the DNA molecule winding around histones to form nucleosomes, which are connected to each other by DNA and may resemble Figure 5.12 on page 216 of the student textbook. 13. a. here is no set relationship between the complexity
of an organism (number of genes in an organism) and the total size of its genome. An organism may have an enormous number of base pairs in its genome and very few genes if the bulk of its genome consists of non-coding DNA.
b. Comparing the genomes of the two organisms would show what genes they have in common, and would indicate their evolutionary relationship—how closely or distantly related they are.
14. A mutation in a protein-coding region would not necessarily be more detrimental than a mutation in a non-coding region since the latter may contain regulatory sequences (i.e., regions that can inluence the production of proteins and RNA molecules). In addition, since multiple codons exist for a given amino acid, a mutation in the protein-coding region of DNA may not alter the protein sequence.
Answers to Section 5.2 Review Questions
(Student textbook page 229)1. Daughter cells must have the same genetic information as parent cells.
2. a. Flowcharts should include a summary of
experimental steps and results shown in Figure 5.16 on page 221 of the student textbook.
b. If DNA replication was conservative, only two bands would appear following one round of replication: one band of 14N-only DNA (newly
synthesized DNA), and one band of 15N-only DNA (old parental DNA). Diagrams should show two distinct bands, one labelled “light (14N) and the
other labelled heavy (15N).
3. • Initiation—Replication begins at the replication origin. Helicases bind to the DNA at each replication origin. he helicases cleave and unravel a section of the original double helix, creating Y-shaped areas (replication forks) at the end of the unwound areas, which form a replication bubble. Single-strand binding proteins stabilize the separated strands. hese single strands serve as templates for the semi-conservative replication of DNA.
• Elongation—New DNA strands are produced when DNA polymerase inserts into the replication bubble. A primase synthesizes an RNA primer that serves as the starting point of new nucleotide attachment
by DNA polymerase. DNA polymerase can only synthesize the new nucleotide chain in the 5′ to 3′
direction. As a result, one strand (the leading strand) is replicated continuously in the 5′ to 3′ direction, in the same direction that the replication fork is moving. he other strand, known as the lagging strand, is replicated in short segments, still in the 5′ to 3′
direction, but away from the replication fork. hese fragments, called Okazaki fragments, are joined together by DNA ligase.
• Termination—When replication is complete, the two new DNA molecules separate from one another and the replication machine is dismantled. Each new molecule of DNA contains one parent strand and one new strand.
4. Early development is a very rapid process, and many key molecules are produced during this time. herefore, it is expected that more replication origins would be present in developing embryo cells.
5. A replication bubble is formed as the DNA double helix unwinds during initiation. he replication forks are the Y-shaped regions of the replication bubble, and move along the DNA in opposite directions as replication proceeds. Diagrams should resemble the third portion of Figure 5.17 on page 223 of the student textbook, with labels on the replication bubble, replication fork, and the double-headed arrow showing the direction(s) of unwinding.
6. Diagrams should illustrate continuous DNA synthesis on the leading strand and discontinuous DNA synthesis on the lagging strand. Diagrams may resemble a simpliied version of Figure 5.19 on page 224 of the student textbook with labels for leading strand, lagging strand, Okazaki fragments, RNA primer, DNA polymerase, DNA ligase, parent DNA, and directionality of strands.
7. An RNA primer is necessary for DNA synthesis on the lagging strand. he primer provides a free 3′-hydroxyl end, which DNA polymerase can extend by adding new nucleotides.
8. DNA polymerase adds new nucleotides to the 3′ end of a growing chain during replication. DNA polymerase also proofreads newly formed base pairs and replaces any nucleotides that have been incorrectly added. 9. a. No RNA primer would be synthesized. herefore,
synthesis of the lagging strand cannot be initiated. b. Okazaki fragments on the lagging strand cannot be
joined together.
10. A—he semi-conservative model states that each new molecule of DNA would contain one strand of original parent DNA and one new strand of daughter DNA. B—he dispersive model states that new molecules of DNA would be hybrids containing a mixture of old parent DNA and new daughter DNA strands.
C—he conservative model states that one molecule of DNA would contain two new daughter DNA strands, and the other molecule of DNA would contain the original parent DNA strands.
11. Graphic organizers should include:
• Prokaryotes Only—rate of replication is faster compared to eukaryotes; ive DNA polymerases have been identiied in prokaryotes; circular chromosome of prokaryotes have a single replication origin • Prokaryotes and Eukaryotes—require replication
origins; have 5′–3′ elongation; have continuous synthesis on the leading strand, and discontinuous synthesis on the lagging strand; require a primer for Okazaki fragments on the lagging strand; require the use of DNA polymerase enzymes
• Eukaryotes Only—rate of replication is slower compared to prokaryotes due to complicated enzymes complexes and proofreading mechanisms; 13 DNA polymerase enzymes have been identiied in eukaryotes; linear chromosome has multiple replication origins; presence of telomeres due to linear nature of eukaryotic chromosome
Answers to Chapter 5 Review Questions
(Student textbook pages 235–9) 1. d2. b
3. e
4. b
5. c
6. c
7. b
8. d
9. b
10. a
11. d
12. d
13. a
14. a
15. a. While studying DNA in the early 1900s, Phoebus Levene reported that the nucleotides were present in equal amounts, and that they appeared in chains in a constant and repeated sequence of nitrogen bases. herefore, most scientists thought that the great variety of proteins was an important factor, and must be the hereditary material. Scientists assumed that the molecular structure of DNA was just too simple to provide the great variation in inherited traits.
b. Oswald Avery, Colin MacLeod, and Maclyn McCarty conducted a series of experiments and discovered: • When they treated heat-killed pathogenic bacteria
with a protein-destroying enzyme, transformation still occurred.
• When they treated heat-killed pathogenic bacteria with a DNA-destroying enzyme, transformation did not occur.
hese results provided evidence that genetic information was carried on DNA.
16. Franklin’s X-ray difraction images showed that DNA had a helical structure, with two regularly repeating patterns. She also concluded that the sugar-phosphate backbone was located on the outside, and the nitrogen-containing bases protruded inward. Watson and Crick used these observations to construct the three-dimensional model of DNA.
17. a. Each body cell produces two daughter cells from itself, and one of the two strands could go to each daughter cell.
b. With bases on the outside, the DNA would not be uniform width throughout, which all evidence indicated. Also, the weak hydrogen bonds between the nitrogen bases could be broken easily. he bonds between the sugar and phosphate portions of the nucleotides are much stronger.
c. From Franklin’s X-ray photographs, they reasoned that DNA was twisted into a spiral, or helix. Since the spiral consisted of two strands wound around each other, they called it a double helix.
18. Eukaryotic DNA is compacted in the nucleus through diferent levels of organization. DNA associates with histones to form nucleosomes. It can be further compacted by the coiling of nucleosomes to produce 30 nm ibres. Additional compacting is achieved through the formation of loop domains of the 30 nm ibre on a protein scafold. his scafold can condense further through folding.
20. Hydrogen bonds between the bases hold the two strands of DNA together.
21. he two strands of a DNA molecule are antiparallel since each strand has directionality. At each end of the DNA molecule, the 5′ end of one strand is across from the 3′ end of the complementary strand.
22. Since nitrogen is a component of DNA, it would be incorporated into newly synthesized strands of DNA. Two diferent isotopes of nitrogen were used to distinguish between the original parental strand and the newly synthesized daughter strand.
Furthermore, having a “light” form (14N) and a “heavy” form (15N) of nitrogen allowed the separation
of diferent DNA strands based on the amount of isotope present in the newly synthesized DNA. DNA with more 15N would be denser than DNA with
14N, and therefore could be separated by centrifuge
and visualized.
23. Producing exact copies ensures that when a cell divides, the ofspring cells will receive the same genetic information as the parent cell.
24. An RNA primer is required for discontinuous synthesis of DNA on the lagging strand. he RNA primer provides a free 3′ hydroxyl end from which DNA polymerase can add nucleotides.
25. he replication machine consists of the complex of proteins and DNA that interact at the replication fork. hese proteins include DNA polymerase, an enzyme that joins nucleotides together to create a complementary strand of DNA (elongation); DNA ligase, an enzyme that joins Okazaki fragments together; primase, an enzyme that constructs the RNA primer needed for replication to begin; helicases, a group of enzymes that cleave and unravel a segment of the double helix to enable replication; and single-strand binding proteins, which help stabilize the unwound strands.
26. Having multiple origins of replication increases the speed of replication. Instead of starting at one end and inish at another, having multiple start points increases the eiciency of replication.
27. DNA polymerase has a proofreading mechanism, which identiies and excises an incorrect nucleotide, and then inserts the correct nucleotide. Another mechanism of error correction is mismatch repair, where a group of enzymes can detect deformities in the newly synthesized strand of DNA caused by mispairing of nucleotides. his group of enzymes excises the mispaired nucleotides and inserts the correctly paired nucleotide. Errors that are not corrected before cell
division become mutations in the genome, which are passed on to daughter cells once cell division occurs.
28. Telomeres are present to ensure that important genetic information is not lost during replication of linear eukaryotic DNA.
29. Nucleotides can come fully-formed from a variety of food sources. Nucleotides can also be formed by our bodies using components that come from our diet (i.e., sugar, phosphates, nitrogen).
30. Diferent tissues all develop from the same fertilized egg cell (zygote). While the tissues have the same genes, only those genes necessary for a speciic tissue’s functions are active.
31. Sample answer: here are some similarities. However, DNA “words” are limited to sequences of amino acids. Each section of code has only “one meaning,” resulting in one speciic protein. his does not compare to the arrangement of letters in a language, which results in words that can have a great variety of meanings.
32. a. 5′-GATGTACAG-3′
b. 5′-ATCAGCGAT-3′
c. 5′-AATACGCCG-3′
33. G = 16%, T = 34%
34. Sample B is the viral DNA because the percentages of adenine and thymine are not the same. Similarly, the percentages of guanine and cytosine are not the same, as they are in sample A, which shows complementary base pairing of these respective bases. Complementary base pairing does not occur in a single-stranded DNA virus.
35. a. Sample answer: he small fragments could be Okazaki fragments that were not joined together properly. his could be due to a lack of ligase, a mutant ligase, or a mutation in the ligase gene that produces a mutant ligase enzyme.
b. Experimental design should include the experimental setup, proper controls (i.e., one reaction tube with cells cultured without ligase, and another with ligase), and expected results.
36. A similar base composition does not necessarily mean that the DNA sequences are similar (i.e., the order of the nucleotides in the DNA sequence are not necessarily the same in both organisms).
of eukaryotic DNA organization with labels for DNA, histones, nucleosomes, and linker DNA (refer to Figure 5.12 on page 216 of the student textbook). An accompanying diagram should illustrate micrococcal nuclease cutting linker DNA between nucleosomes. 38. Answers and diagrams should include:
• If replication was continuous in a 5′ to 3′ direction, the two DNA strands would not be antiparallel and would instead be parallel to each other.
• If continuous replication was able to occur in both 5′ to 3′ and 3′ to 5′ directions, then primase, RNA primers, Okazaki fragments, and DNA ligase would not be necessary for synthesis on the lagging strand. Additionally, telomeres would not be necessary since there would be complete synthesis of the lagging strand of eukaryotic DNA.
39. he weak hydrogen bonds in DNA break easily, making it easier for the two strands in the molecule to separate during replication. he strong covalent bonds ensure that the sequence of nucleotides remains ixed in each strand.
40. Radioactive phosphorus (32P) would label newly-synthesized DNA strands. Diagrams should resemble the semi-conservative model of replication shown in Figure 5.15 on page 220 of the student textbook, where the 32P-labelled strands would correspond to the newly synthesized daughter strands. Ater two rounds of replication, both strands of new DNA double helix should incorporate 32P.
41. 5′-UGACU-3′
42. DNA polymerases are expected to be more active in continuously dividing skin cells, since DNA replication would occur more oten. Heart muscle cells do not divide as frequently as skin cells, and therefore DNA replication (and DNA polymerase activity) occurs less oten.
43. Answers could include targeting bacterial-speciic topoisomerases. hese enzymes are essential for supercoiling and DNA replication, which are both required for the survival of bacterial cells. Since this is a bacteria-speciic target, the side efects of this drug on eukaryotic cells could be reduced.
44. A defect in DNA helicase could result in delayed unwinding of the DNA double helix. herefore, the initiation of DNA replication would not occur eiciently, which could lead to DNA instability and cell death.
45. Primase would no longer be needed to synthesize a RNA primer on the lagging strand, since a 3′ hydroxyl end would already exist. Ligase would not be necessary since Okazaki fragments would not be synthesized on the lagging strand.
46. Concept maps should include information on two or more experiments that addressed a similar hypothesis (i.e., the transforming principle experiments performed by Griith and Avery). he experimental approaches and results for each experiment should be outlined in the concept map, in addition to how the diferent experiments are related to one another (i.e., did the result from one experiment help direct subsequent experiments?). Using a variety of experimental approaches provides conidence and conirmation of results.
47. Timelines should be a chronological order of contributions from the scientists presented in the chapter: Miescher; Levene; Griith; Avery, MacLeod, and McCarty; Hershey and Chase; Chargaf; Pauling; Franklin; Watson and Crick.
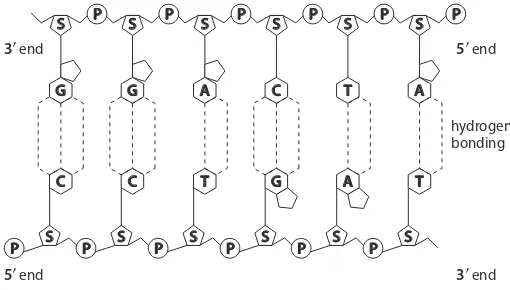
48. Sample diagram:
P S
C G
C G
T A
A T
T A
G C P
S S P S P S P S P
P S P S P S P S P S P S
5′ end 3′ end
3′ end hydrogen bonding
5′ end
49. Graphic organizers should include:
• Prokaryotes Only—rate of replication is faster compared to eukaryotes; ive DNA polymerases have been identiied in prokaryotes; circular chromosome of prokaryotes have a single replication origin. • Prokaryotes and Eukaryotes—require replication
origins; have 5′-3′ elongation; have continuous synthesis on the leading strand, and discontinuous synthesis on the lagging strand; require a primer for Okazaki fragments on the lagging strand; require the use of DNA polymerase enzymes
50. Diagrams should resemble the bottom illustration in Figure 5.16 on page 221 of the student textbook, with the irst set of parental and new strands labelled irst round, and the next set labelled second round. 51. Flowcharts should include:
• Initiation—Helicase enzymes unwind the DNA double helix to separate it into two strands. A replication bubble and replication forks are formed when single-strand binding proteins stabilize the separated strands. Topoisomerase enzymes help to relieve the strain on DNA caused by unwinding. • Elongation—New DNA strands are synthesized
by joining free nucleotides together. his is catalyzed by DNA polymerase, which synthesizes the new strands that are complementary to the parental strand. Synthesis of the new DNA strands occurs continuously on the leading strand, and discontinuously on the lagging strand.
• Termination—he two new DNA molecules, each composed of one parental strand and one new daughter strand, separate from one another.
52. Diagrams should resemble Figure 5.17 on page 223 of the student textbook, with labels for replication bubble, replication forks (each end of the bubble), and the double-headed arrow showing the direction(s) of unwinding.
53. DNA can only be synthesized in the 5′ to 3′ direction. Diagram should therefore illustrate continuous DNA synthesis on the leading strand and discontinuous DNA synthesis on the lagging strand. Diagram may
resemble a simpliied version of Figure 5.19 on page 224 of the student textbook with labels for leading strand, lagging strand, Okazaki fragments, RNA primer, DNA polymerase, DNA ligase, parent DNA, and directionality of strands.
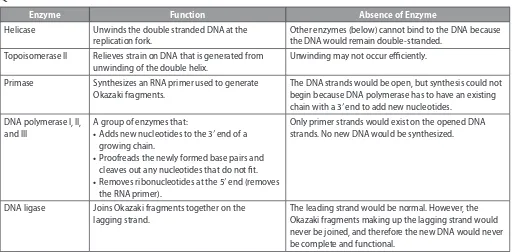
54. See Table below.
55. Journal entries should be written in the irst person and summarize the knowledge of heredity related to his or her research, using scientiic terminology. Challenges could include a lack of research facilities, lack of inancial support, the shortcomings of the current technology, lack of consensus among research colleagues, or lack of support for the work in the broader scientiic community. houghts of future signiicance of the research may relect later discoveries.
56. Articles should summarize Watson and Crick’s indings, including their model of DNA and include how this discovery would afect the general public (i.e., advances in medicine and genetics, ethical issues and concerns). he article should be written in language that is aimed at the general public and is appropriate for the time period.
57. Concept maps should illustrate the diferent levels of organization of eukaryotic DNA. Answers may resemble Figure 5.12 on student textbook page 216, with the addition of a nucleotide label above the DNA molecule, nucleosome at the structure illustrating DNA wrapped around histones, and chromatin on all of the non-condensed forms of genetic material.
Enzyme Function Absence of Enzyme
Helicase Unwinds the double stranded DNA at the replication fork.
Other enzymes (below) cannot bind to the DNA because the DNA would remain double-stranded.
Topoisomerase II Relieves strain on DNA that is generated from unwinding of the double helix.
Unwinding may not occur eiciently.
Primase Synthesizes an RNA primer used to generate Okazaki fragments.
The DNA strands would be open, but synthesis could not begin because DNA polymerase has to have an existing chain with a 3′ end to add new nucleotides.
DNA polymerase I, II, and III
A group of enzymes that:
• Adds new nucleotides to the 3′ end of a growing chain.
• Proofreads the newly formed base pairs and cleaves out any nucleotides that do not it. • Removes ribonucleotides at the 5′ end (removes
the RNA primer).
Only primer strands would exist on the opened DNA strands. No new DNA would be synthesized.
DNA ligase Joins Okazaki fragments together on the lagging strand.
The leading strand would be normal. However, the Okazaki fragments making up the lagging strand would never be joined, and therefore the new DNA would never be complete and functional.
58. his would leave replication errors (i.e., mispairing of bases) uncorrected. hese errors would then become mutations in the genome, which are then passed onto daughter cells once cell division occurs.
59. he graphic organizer should efectively represent the points outlined in the Chapter 5 summary on page 234 of the student textbook.
60. Answers may include:
• Human disease—Many genes that are implicated in human disease have parallel versions in model organisms (i.e., yeast, mice, fruit lies), where they can be studied easily in various experimental settings. • Gene function—Studying parallel human genes
in model organisms may also provide insight into gene function.
• Evolutionary biology—Comparative genomics also allows researchers to study which regions of a genome have been conserved amongst diferent species. hese conserved regions are thought to be essential and important regions of the genome. Likewise, divergent regions may confer species-speciic function and contribute to morphological and functional changes.
61. Answers should include speciic examples of how the chosen scientist’s research would beneit the scientiic research community and the general public. Social, legal, and ethical implications may be addressed. he paragraph should be convincing, as this is an example of writing that would be included in a grant application.
62. a. When the last RNA primer from the lagging strand is degraded in linear DNA, the gap that remains is unable to be illed since there is no adjacent fragment where nucleotides can be added. herefore, the new DNA molecule will be shorter than the parent DNA molecule. Since telomeres are at the ends of eukaryotic chromosomes, they are the sequences which become shorter ater each round of replication.
b. Telomerase is an enzyme that synthesizes telomeres and replaces sequences that have been lost. In childhood, telomerase activity in cells is high. As people age, the activity of telomerase decreases, which can result in shorter telomeres and therefore shorter chromosomes. his may lead to loss of important coding information. Certain lifestyle factors may inluence telomere length based on its efects on telomerase activity. For example, smoking may cause accelerated symptoms of aging due to
decreasing telomerase activity. Exercise, on the other hand, may delay the symptoms of aging since it increases telomerase activity.
63. Answers may include:
• If there was a method to increase telomerase activity, who would have access to “fountain of youth” technology?
• Should we be interfering with the natural aging process?
• Why would certain individuals want to delay aging? • What are the possible beneits and risks of being able
to delay aging?
64. a. Telomerase activity may be higher in cancer cells since these cells divide rapidly.
b. Telomerase is present and active in normal cells. Inhibiting telomerase activity as a potential cancer therapy could therefore cause damage to normal cells (i.e., shortened or lack of telomeres, which would lead to more rapid shortening of chromosomes).
65. a. Students’ opinions should be supported with examples.
b. Issues may addressed include:
• How would appropriate credit be determined? • Should contributions be based on the percentage of
work performed for the study or for the analysis? • Should primary credit go to the scientist who
proposed the hypothesis, or should it go to the scientist who actually carried out the experiment (i.e., did the “bench work”)?
66. Answers may be based on cultural, religious, or family values. Accept any reasoned argument.
67. Answers may include: • DNA sequencing
• Genetic screening for diseases • herapeutic gene targets
• Gene expression (RNA and protein) • DNA mutations
68. his required a rethinking of many ideas that formed the basis of other ideas. he lower number of genes may mean that
• the structures function diferently than anticipated • the number of number genes does not determine an
organism’s complexity
• there are a large number and sizes of introns • non-coding regions act as regulatory sequences • genes work in combinations (groups) to perform
69. In Linus Pauling’s model, DNA replication would have to occur without nitrogen base pairing. Accept any well-reasoned answer.
70. Anti-viral drugs speciically target viral DNA
polymerase to interfere with viral replication. Speciicity is also required to ensure eukaryotic DNA polymerase activity in not adversely afected.
71. a. Because of the shared features/structure of their DNA.
b. Graphic organizers (such as a table) should include the advantages and disadvantages of using the chosen organism and speciic examples of signiicant research indings that were obtained using that organism. c. Ethical issues will vary but may include the harm
done to the animal either by being kept in captivity or by the experimental process.
72. a. Mispairing of bases. hymine should be paired with adenine, and cytosine should be paired with guanine.
b. Mispairing of bases occurs during replication and may be due to lexibility in the structure of DNA. c. Mismatch repair can correct this error. A group of
mismatch repair enzymes recognizes deformities that are caused by mispairing of bases. hese enzymes then excise the incorrect nucleotide and insert the correct nucleotide. DNA polymerases can also correct this error by excising the incorrect nucleotide in a newly synthesized strand, and adding the correctly paired nucleotide.
Answers to Chapter 5 Self-Assessment Questions
(Student textbook pages 240–1)1. b
2. c
3. e
4. c
5. a
6. e
7. c
8. a
9. e
10. b
11. C = 19%, G = 19%, T = 31%
12. a. Two diferent radioactive isotopes were used to trace each type of molecule. One sample of T2 virus was tagged with radioactive phosphorus (32P), since
phosphorus is present in DNA but not in protein. he other sample of T2 virus was tagged with radioactive sulfur (35S), since sulfur is only found
in the protein coat of the capsid.
b. In one experiment, Hershey and Chase observed that most of the radioactively labelled viral DNA was in bacteria and not in the liquid medium. In a second experiment, they observed that the radioactively labelled viral capsid protein was in the liquid medium and not in the bacteria. hese results demonstrated that viral DNA, not viral protein, enters the bacterial cell. herefore, DNA is the hereditary material.
13. Franklin’s X-ray difraction images showed that DNA has a helical structure with two regularly repeating patterns. She also concluded that the nitrogenous bases were located on the inside of the helical structure, and the sugar-phosphate backbone was located on the outside, facing toward the watery nucleus of the cell.
14. Diagrams should include labels for sugar-phosphate molecules (“handrails”), nucleotide base pairing (“rungs”), and directionality of both strands and resemble the close up part of Figure 5.7 on page 213 of the student textbook.
15. Prokaryotic DNA is double-stranded, circular, and packed in the nucleoid. It is compacted via supercoiling.
Linear eukaryotic DNA is compacted in the nucleus through diferent levels of organization. DNA associates with histones to form nucleosomes. It can be further compacted by the coiling of nucleosomes to produce 30 nm ibres. Additional compacting is achieved through the formation of loop domains of the 30 nm ibre on a protein scafold. his scafold can condense further through folding.
he diferences are due to structure (circular
prokaryotic DNA versus linear eukaryotic DNA) and size. Since the total amount of DNA in eukaryotes is much greater than in prokaryotes, they have greater compacting and levels of organization.
16. Answers should include support for the student’s interpretation of the statement. While protein-coding regions include genes which code for proteins, the non-coding regions have regulatory sequences which can inluence and regulate the production of proteins and RNA molecules.
b. Answers could include forensics, comparative genomics, screening for diseases, or tracing the origin or source of an illness.
c. Opinions should be supported with an explanation that includes evidence of scientiic understanding of the nature of a DNA sequence. Answers may also consider consent under certain circumstances (i.e., genetic screening for disease or identifying lineage). 18. a. Arrow should indicate movement to the let.
b. Okazaki fragment C was made irst. Primase synthesizes an RNA primer, which binds to the parental strand of DNA. DNA polymerase III adds new nucleotides to the free 3′-hydroxyl end of the primer. his newly synthesized fragment is an Okazaki fragment.
c. Okazaki fragments are necessary for the
discontinuous synthesis of the lagging strand during DNA replication. Once the Okazaki fragments are made, DNA polymerase I removes the RNA primer and the Okazaki fragments are joined together by DNA ligase to form a complete lagging strand.
19. DNA has weak hydrogen bonds, which exist between the nucleotides on opposite strands. hese weak hydrogen bonds break easily, making it easier for the two strands to separate during replication. he strong covalent bonds that exist on the sugar-phosphate backbone ensure that the sequence of nucleotides remains ixed in each strand, since these bonds are not easily broken.
20. Diagrams should resemble Figure 5.17 on page 223 of the student textbook, with labels identifying the replication bubble, replication forks at each end of the bubble, and the double-headed arrow showing the direction of unwinding.
21. Answers should include the information summarized in Table 5.2 on page 224 of the student textbook, which lists the important proteins involved in DNA replication and their functions (helicase, primase, single-strand-binding protein, topoisomerase II, DNA polymerase I, II, and III, and DNA ligase).
22. A person without DNA polymerase would not exist, because DNA replication could not occur and cells would not be able to divide and survive.
23. Mutations in Mut genes could lead to an inability to correct errors in replication, such as mispairing of bases. Uncorrected errors become mutations in the genome, which are then passed on to daughter cells once cell division occurs.
24. Telomerase activity decreases as we get older, which results in the shortening of telomeres in somatic cells. his means that the age of an organism is relected in the length of telomeres.
25. Venn diagrams should include:
• Prokaryotes Only—rate of replication is faster; ive DNA polymerases identiied; circular chromosome of prokaryotes have a single replication origin • Prokayotes and Eukaryotes—require replication
origins; have 5′-3′ elongation; have continuous synthesis on the leading strand, and discontinuous synthesis on the lagging strand; require a primer for Okazaki fragments on the lagging strand; require the use of DNA polymerase enzymes
• Eukaryotes Only—rate of replication is slower due to complicated enzymes complexes and proofreading mechanisms; 13 DNA polymerase enzymes identiied; linear chromosome has multiple replication origins; presence of telomeres due to linear nature of eukaryotic chromosome
Chapter 6 Gene Expression
Answers to Learning Check Questions
(Student textbook page 246)1. he black urine phenotype shown to be caused by a recessive inheritance factor (gene) that caused that production of a defective enzyme (protein).
2. Answers could use Figure 6.1 on page 245 of the student textbook as a guideline. he results of the Beadle and Tatum experiment showed that a single gene produces one enzyme (one-gene/one-enzyme hypothesis). his was later modiied to the one-gene/one-polypeptide hypothesis since not all proteins are enzymes. 3. RNA is found in the nucleus and cytoplasm; the
concentration of RNA in the cytoplasm is correlated with protein production; RNA is synthesized in the nucleus and transported to the cytoplasm.
4. Jacob and colleagues saw that bacteria infected with a virus had a newly synthesized virus-speciic RNA molecule. his RNA molecule associated with bacterial ribosomes, which are the sites of protein production. herefore, the RNA molecule carried the genetic information for the production of a viral protein. 5. Having DNA continually transport itself could increase
the likelihood of damage to the DNA. Multiple steps in gene expression provide many opportunities for regulation. his allows the cell to have increased control over protein synthesis.
(Student textbook page 254)
7. Answers could include: DNA is the genetic material that controls protein synthesis, whereas RNA helps DNA and is involved in protein synthesis. DNA has the sugar called deoxyribose whereas RNA has ribose sugar. he bases in DNA are adenine, guanine, thymine, and cytosine; the bases in RNA include adenine, guanine, cytosine, and uracil. DNA is a double-stranded helix, and RNA is single stranded without a helix.
8. Answers could include information in Table 6.3 on page 252 of the student textbook: mRNA is the template for translation, while tRNA and rRNA are involved in the translation of mRNA.
9. Initiation—Transcriptional machinery is assembled on the sense strand. RNA polymerase binds to the promoter region of the sense strand.
Elongation—RNA polymerase synthesizes a strand of mRNA that is complementary to the sense strand of DNA.
Termination—RNA polymerase detaches from the DNA strand when it reaches a stop signal. he mRNA strand is release and the DNA double helix re-forms.
10. To the cytoplasm, for translation
11. During the elongation phase of transcription, a second RNA polymerase complex can bind to the promoter region immediately ater the previous RNA polymerase complex starts moving along the DNA. herefore, multiple strands of mRNA can be synthesized from one gene at a time. his is advantageous for the cell since having more mRNA leads to more protein that can be produced in a given amount of time.
12. Errors in transcription would be less damaging than errors in DNA replication. An error in transcription would results in an error in one protein molecule, while an unrepaired error in DNA replication would cause a change in the genetic makeup of an organism.
(Student textbook page 260)
13. Answers may be based on Table 6.4 on page 257 of the student textbook and should include:
• mRNA—contains genetic information which determines a protein’s amino acid sequence • tRNA—a molecule that links mRNA codons with
their corresponding amino acid
• ribosomes—a structure composed of rRNA and proteins which provides the site for protein synthesis • translation factors—accessory proteins which are
necessary for each stage of translation
14. A polyribosome is a complex composed of multiple ribosomes along a strand of mRNA. It can produce many copies of a protein at the same time.
15. Translation consumes large amounts of energy since many molecules (i.e., proteins and nuclei acid components) must be synthesized and assembled. Energy is also needed to form peptide bonds that exist between the amino acids in proteins. his energy is provided by mitochondria, which supply the cell with most of its energy.
16. Translation is initiated when initiation factors assemble the translation components. he small ribosomal subunit attaches to the mRNA near the start codon (AUG). he initiator tRNA (with anticodon UAC) binds to the start codon. he large ribosomal subunit then joins to form the complete ribosome. Translation is terminated when a stop codon on the mRNA is reached. his causes the polypeptide and the translation machinery to separate. he polypeptide is then cleaved from the last tRNA by a release factor.
17. Answers could be based on Figure 6.14 on page 259 of the student textbook.
18. he antibiotic would prevent the initiation of translation of bacterial proteins. herefore, the bacterial cells would not survive.
(Student textbook page 265)
19. Mutations that occur in reproductive cells can be passed on to future generations. Mutations which occur in somatic cells do not get passed on from one generation to another.
20. No, since the deletion of nucleotides is divisible by three.
21. A silent mutation has no efect on the amino acids of a protein, and therefore would be the least harmful to an organism. Missense mutations cause alterations in the amino acids of a protein, and nonsense mutations results in a shortened protein due to a premature stop codon.
22. Single-gene mutations involve changes to the nucleotide sequence of one gene. Chromosomal mutations involve changes in chromosomes, and therefore may afect many genes.
23. Spontaneous mutations may arise during DNA replication if there is incorrect base pairing by DNA polymerase. DNA transposition can also cause
24. UV radiation can lead to a mutation by covalently linking thymines to form a thymine dimer. A speciic mechanism for repairing thymine dimers is photorepair, where an enzyme speciically recognizes and cleaves thymine dimers. A non-speciic
mechanism for repairing UV damage is excision repair, where a group of excision repair enzymes identify and correct many diferent types of DNA damage.
(Student textbook page 269)
25. Constitutive genes code for proteins that needed for cell survival. herefore, they are always active and are expressed at constant levels.
26. he regulation of genes allows for cell specialization. Gene expression is controlled so that only certain genes are active at certain levels and at certain times in each cell type. herefore, each cell type can have its own set of proteins.
27. Many genes in bacteria are clustered together in operons and are therefore under transcriptional control of a single promoter. his type of transcriptional control is energy eicient since these genes are transcribed together into a single polycistronic mRNA, rather than individual mRNA molecules. Individual proteins are then synthesized from the polycistronic mRNA.
28. Answer could be based on Figure 6.23 on page 267 of the student textbook. Diagram should include the regulatory region, promoter region, operator, and the coding region.
29. he lac operon is inducible since the transcription of genes is induced when lactose is present. In the
trp operon, the genes are actively transcribed until a repressor binds to inhibit transcription.
30. A common analogy used is the regulation of temperature by a thermostat, since the feedback mechanism is similar (i.e., at low temperatures, the thermostat turns on to increase the temperature; at high temperatures, the thermostat turns of to decrease the temperature).
Answers to Caption Questions
Figure 6.1 (Student textbook page 245): Growth would occur ater each addition of intermediates since glutamate is the irst step of the arginine pathway.
Figure 6.3 (Student textbook page 249): he drugs will also inhibit translation, since it follows transcription.
Figure 6.8 (Student textbook page 253): he increasing length of the mRNA strands indicates the direction of transcription.
Figure 6.11 (Student textbook page 257): he genes that code for tRNA produce tRNA molecules, which are not translated into proteins.
Figure 6.16 (Student textbook page 261): Cysteine
Figure 6.18 (Student textbook page 263): A silent mutation occurs when a nucleotide substitution has no efect on the polypeptide sequence (i.e., ACU to ACC)
Figure 6.24 (Student textbook page 268): Both operons have the same basic structure. he lac operon is inactive unless lactose is present. he trp operon is normally active, but becomes inactive if enough tryptophan is made.
Answers to Section 6.1 Review Questions
(Student textbook page 250)1. While Garrod’s indings suggested a link between genes and proteins, it was Beadle and Tatum who found experimental evidence of the relationship between genes and proteins.
2. a. he arg mutants would be able to grow since all the required amino acids for growth are provided in the medium.
b. It would be impossible to determine what enzyme in the arginine pathway is afected by each mutation since all of the mutants would be able to grow on the complete medium (i.e., the medium that is supplemented with all the required amino acids for growth). herefore, these results would not be able to show that one gene codes for one enzyme. 3. Questions may include:
• Why was the one-gene/one-enzyme hypothesis updated to the one-gene/one-polypeptide hypothesis? • Is the one-gene/one-polypeptide hypothesis an
accurate description of gene expression? Explain your answer.
• If you could update the one-gene/one-polypeptide hypothesis, what changes would you make? Explain your answer.
4. Jacob and colleagues showed that messenger RNA (mRNA) acts as a “genetic messenger.” mRNA is synthesized from the DNA of genes and carries information to produce a protein.
5. he triplet hypothesis proposes that the genetic code consists of a combination of three nucleotides (codon). 6. a. Answers may resemble Figure 6.2 on page 247 of
the student textbook, showing experimental set-up and results.
7. Answers should include an explanation of why the suggested genetic code system could be used. Genetic codes should be able to code for 20 diferent amino acids, with some duplication. For example, a genetic code of four nucleotides per codon could result in a maximum of 44 or 256 amino acids. his allows for the
20 diferent amino acids, with some duplication. 8. • A redundant genetic code means that more than one
codon can code for the same amino acid.
• A continuous genetic code means that the code reads as a continuous series of three-letter codons without spaces, punctuation, or overlap.
• A universal genetic code means that almost all living things use the same genetic code to build proteins.
9. Because the start codon AUG also codes for methionine.
10. Stop codons, also known as terminator codons, signal the end of polypeptide growth.
11. Answers should indicate that the genetic code is redundant—that is, more than one codon can code for the same amino acid. Only three codons do not code for any amino acid. he redundancy in the genetic code is extremely valuable to the organism. For example, if a mutation occurs to the DNA, causing an AAT sequence to become AAC, the mRNA codon transcribed by the original DNA is UUA while the mutated DNA transcribes a UUG codon. he mutation, however, will not harm the organism because these two codons both correspond to leucine.
12. a. he central dogma of genetics.
b. Genetic information lows from DNA to RNA to protein.
13. Almost all living organisms have the same genetic code. Everyone has his or her own unique set of nucleotides (and therefore DNA bases).
14. Amino acid sequence: arg-iso-val-ser-leu
Key: arg = arginine; iso = isoleucine; val = valine; ser = serine; leu = leucine
15. a. he universality of the genetic code refers to the discovery that the same genetic code is used by all living things. his indicates that all living things share similar ancestry. For example, a codon in a fruit ly codes for the same amino acid in humans. b. he universality of the genetic code has signiicant
implications for molecular biology techniques, such as cloning. For example, a gene from one organism can be inserted into a diferent organism to produce the same protein.
Answers to Section 6.2 Review Questions
(Student textbook page 256)1. Each type of RNA has a diferent function in the cell. Some RNA are involved in translation (e.g., tRNA, rRNA), while others act as part of an enzyme (e.g., RNA in RNAses). Answers may refer to examples in Table 6.3 on page 252 of the student textbook, which lists the diferent types of RNA molecules and their functions.
2. Flowcharts should include:
• Initiation—Transcriptional machinery is assembled on the sense strand. RNA polymerase binds to the promoter region of the sense strand.
• Elongation—RNA polymerase synthesizes a strand of mRNA that is complementary to the sense strand of DNA.
• Termination—RNA polymerase detaches from the DNA strand when it reaches a stop signal. he mRNA strand is release and the DNA double helix re-forms. 3. he objective of transcription is to produce an accurate
copy of a small section of genomic DNA in the form of mRNA. he objective of DNA replication is to produce two identical DNA molecules from a parent DNA molecule. While they have deined stages, diferent proteins are used in transcription (e.g., RNA polymerase instead of DNA polymerase).
4. For each gene, only one strand of the DNA molecule is transcribed. he sense strand is also known as the coding strand—the strand that is transcribed. he antisense strand is also known as the non-coding strand—it is not transcribed.
5. A promoter region is a sequence of nucleotides on DNA where RNA polymerase complex binds. hese regions are important for transcription because RNA polymerase must bind to them for transcription to be initiated.
6. Venn diagrams should include:
• DNA polymerase Only—synthesizes two new strands of DNA from template DNA; has proofreading abilities
• DNA and RNA polymerase—work in a 5′-3′
direction; add new nucleotides to the 3′-hydroxyl group of the previous nucleotide
7. a. 5′-TATGATCAC-3′
b. sense strand
c. 5′-UAUGAUCAC-3′
d. tyrosine-aspartate-histidine
8. Sample answer: Because DNA is double stranded, DNA polymerase is able to recognize whether hydrogen bonding is taking place between a base in the newly synthesized strand of DNA and its complement in the original strand. he absence of hydrogen bonding indicates a mismatch between bases, and DNA polymerase excises the incorrect base and inserts the correct one. his proofreading ability reduces the incidence of mutation in the genetic code and the possible translation of a non-functional protein.
9. Protein synthesis cannot occur at the same time as transcription since eukaryotic mRNA must undergo modiication before it can be transported from the nucleus to the cytoplasm.
10. a. 5′ cap, Exon, Exon, Exon, AAA, 3′
b. 5′ cap—a modiied G nucleotide covalently links to the 5′ end; recognized by protein synthesis machinery
Splicing—introns removed and exons are joined together
poly-A tail—a series of A nucleotides covalently linked to the 3′ end; makes mRNA more stable 11. he discrepancy in the number of genes and the
number of proteins in the human proteins may be explained by alternative splicing, where one gene can code for more than one protein. Answers should include a diagram illustrating alternative splicing in eukaryotic genes.
12. Alternative splicing allows for one gene to code for more than one protein. For some genes, all exons are spliced together. In certain cases, only certain exons are spliced together to form mature mRNA.
13. a. he presence of introns allows a gene to produce multiple transcripts by alternative splicing. In some cases introns also contain regulatory sequences to control gene expression.
b. In prokaryotes, transcription and translation can occur at the same time since no post-transcriptional modiications (such as removal of introns)
are needed.
Answers to Section 6.3 Review Questions
(Student textbook page 266)1. See Table 6.4 on page 257 of the student textbook.
2. Diagrams should resemble Figure 6.11A on page 257 of the student textbook. Sample caption: Transfer RNA (tRNA) is a molecule that links codons on mRNA to the corresponding amino acid during protein synthesis. Each tRNA molecule contains an anticodon loop, which has complementary base pairs to a speciic mRNA codon, and an acceptor step, which has the corresponding amino acid.
3.
Codon (mRNA) Anticodon (tRNA) Amino Acid
GCC 3′-CGG-5′ alanine
UUU 3′-AAA-5′ lysine
AAC 3′-UUG-5′ asparagines
4. Ribosomes are the sites of protein synthesis. Each ribosome is composed of two sub-units: a small ribosomal sub-unit and a large ribosomal sub-unit. he small ribosomal sub-unit has a binding site for mRNA, while the large ribosomal sub-unit contains the three binding sites for tRNAs.
5. Translation requires large amounts of energy since many molecules must be synthesized and assembled during this process. Energy is also needed to form peptide bonds that exist between the amino acids in proteins.
6. Answers should resemble Figure 6.14 on page 259 of the student textbook, showing one amino acid being added at a time to the growing polypeptide chain. 7. his mutation will not be passed to future ofspring.
Mutations in somatic cells, such as skin cells, do not get passed from one generation to another. Only mutations in reproductive cells will get passed to future generations.
8. Single-gene mutations involve changes in the nucleotide sequence of one gene. Chromosomal mutations involve changes in chromosomes, and involve many genes.
10. Physical mutagens such as X-rays and UV radiation cause mutations by physically changing the structure of DNA.
Chemical mutagens such as nitrites, gasoline fumes, and compounds found in cigarette smoke cause mutations by reacting chemically with DNA.
11. Speciic repair mechanisms involve a set of proteins that are tailored to ix certain types of DNA damage. Photorepair is a speciic repair mechanism that repairs thymine dimers caused by UV radiation. Photolyase enzyme recognizes the damage, binds to the dimer, and then excises the dimer by using visible light.
Non-speciic repair mechanisms can ix diferent types of DNA damage. For example, excision repair involves removing a damaged region of DNA and replacing it with the correct sequence.
12. a. GUU-ACC-UGU-UAU-U; frameshit mutation b. valine-threonine-cysteine-tyrosine
13. a. 5′-AUGAAUGAGCAGUUGGAA-3′
methionine-asparagine-glutamate-glutamine-leucine-glutamate
b. 5′-AUGAAUGAGCAGUAGGAA-3′
methionine-asparagine-glutamate-glutamine c. nonsense mutation
14. a. DNA damage caused by UV radiation is not repaired to due defective photorepair mechanisms. his leads to an accumulation of harmful mutations that could lead to cancer.
b. Individuals with XP would need to avoid UV radiation by protecting themselves from sunlight.
Answers to Section 6.4 Review Questions
(Student textbook page 272)1. Gene regulation allows for cell speciicity. Gene expression is controlled so that only certain genes are active at certain levels and at certain times in each cell type. herefore, each cell type can be specialized and have its own set of proteins.
2. Constitutive genes include genes that code for proteins that are required at constant levels for cell survival and maintenance. Answers may include genes involved in cell cycle regulation, DNA replication, transcription, and translation.
3. Certain genes may only be required at certain times, such as genes involved in early development. If these genes were always active, they may have unwanted side efects on cells, since they are not needed in later development.
4. a. An operon is a cluster of genes that are grouped together in a region that is under the control of a single promoter. An operator is a DNA sequence to which a protein binds to regulate transcription. b. Activators and repressors alter levels of transcription
by interacting with RNA polymerase, transcription factors, and enhancers. he activator enhances the level of transcription, whereas the repressor inhibits the level.
5. Diagrams should resembled Figure 6.23 on page 267 of the student textbook.
6. a. Allolactose binds to the repressor so that the repressor can’t bind to the operator. herefore, transcription of genes proceeds to produce the required enzymes.
b. Lac repressor binds to the operator and prevents RNA polymerase from binding to the promoter, inhibiting transcription of genes.
c. Transcription of genes proceeds, but at a slower rate due to lack of activator protein.
d. Transcription of genes is inhibited.
7. he lac operon is inducible since the transcription of genes is induced when lactose is present. In the trp operon, the genes are actively transcribed and only become inactive if enough tryptophan is made. his causes the binding of a repressor to inhibit transcription.
8. Gene regulation is more complex in eukaryotes than in prokaryotes. In eukaryotes, gene regulation occurs at ive levels: pre-transcriptional, transcription, post-transcription, translational, and post-translational. In eukaryotes, gene regulation occurs at three levels: transcriptional, translational, and post-translational. he most common level of regulation in prokaryotes is at the transcriptional level, where genes are regulated through operons.
9. Constitutive genes would more likely be in less condensed areas since they need to be constantly transcribed. Being in less condensed areas of