Supplemental Material
Pavelites, Bash, Gao and MacKerell, Jr. ÒA Molecular Mechanics Force Field for NAD+,
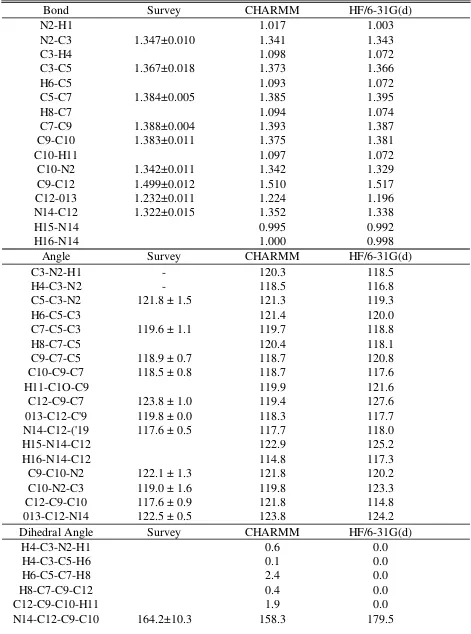
Table S1. Average crystal, ab initio and empirical geometries for NIC+. Bond lengths are given in and angles in given in degrees.
Bond Survey CHARMM HF/6-31G(d)
N2-H1 1.017 1.003
N2-C3 1.347±0.010 1.341 1.343
C3-H4 1.098 1.072
C3-C5 1.367±0.018 1.373 1.366
H6-C5 1.093 1.072
C5-C7 1.384±0.005 1.385 1.395
H8-C7 1.094 1.074
C7-C9 1.388±0.004 1.393 1.387
C9-C10 1.383±0.011 1.375 1.381
C10-H11 1.097 1.072
C10-N2 1.342±0.011 1.342 1.329
C9-C12 1.499±0.012 1.510 1.517
C12-013 1.232±0.011 1.224 1.196
N14-C12 1.322±0.015 1.352 1.338
H15-N14 0.995 0.992
H16-N14 1.000 0.998
Angle Survey CHARMM HF/6-31G(d)
C3-N2-H1 - 120.3 118.5
H4-C3-N2 - 118.5 116.8
C5-C3-N2 121.8 ± 1.5 121.3 119.3
H6-C5-C3 121.4 120.0
C7-C5-C3 119.6 ± 1.1 119.7 118.8
H8-C7-C5 120.4 118.1
C9-C7-C5 118.9 ± 0.7 118.7 120.8
C10-C9-C7 118.5 ± 0.8 118.7 117.6
H11-C1O-C9 119.9 121.6
C12-C9-C7 123.8 ± 1.0 119.4 127.6
013-C12-C'9 119.8 ± 0.0 118.3 117.7
N14-C12-('19 117.6 ± 0.5 117.7 118.0
H15-N14-C12 122.9 125.2
H16-N14-C12 114.8 117.3
C9-C10-N2 122.1 ± 1.3 121.8 120.2
C10-N2-C3 119.0 ± 1.6 119.8 123.3
C12-C9-C10 117.6 ± 0.9 121.8 114.8
013-C12-N14 122.5 ± 0.5 123.8 124.2
Dihedral Angle Survey CHARMM HF/6-31G(d)
H4-C3-N2-H1 0.6 0.0
H4-C3-C5-H6 0.1 0.0
H6-C5-C7-H8 2.4 0.0
H8-C7-C9-C12 0.4 0.0
C12-C9-C10-H11 1.9 0.0
013-C12-C9-C10 8.4±12.2 16.8 0.5
H15-N14-C12-C9 9.6 0.1
H16-N14-C12-C9 172.7 180.0
N2-C3-C5-C7 1.4±1.2 0.0 0.0
C3-C5-C7-C9 1.6±0.9 0.7 0.0
C5-C7-C9-C10 1.2±1.4 1.2 0.0
C7-C9-C10-N2 0.9±2.1 1.1 0.0
C9-C10-N2-C3 1.1±1.3 0.5 0.0
Table S2. NICH crystal, ab initio and empirical geometries. Bond lengths in are given in and angles are given in degrees.
Bond FIXDARa FIXDARb FIXCUK CHARMM HF/6-31G(d)
N2-H1 1.470** 1.467** 1.468** 1.015 0.992
N2-C3 1.369 1.408 1.383 1.371 1.392
C3-H4 0.950 1.095 1.073
C3-C5 1.328 1.328 1.324 1.326 1.320
H6-C5 1.094 1.090 1.075
C5-C7 1.510 1.514 1.479 1.512 1.511
H8-C7 0.950 1.111 1.092
H17-C7 1.572** 1.111 1.090
C7-C9 1.524 1.524 1.527 1.539 1.519
C9-C10 1.352 1.357 1.361 1.353 1.334
C10-H11 1.090 1.094 1.072
C10-N2 1.392 1.354 1.359 1.381 1.365
C9-C12 1.495 1.496 1.456 1.531 1.482
C12-013 1.246 1.238 1.253 1.228 1.206
N14-C12 1.332 1.342 1.334 1.356 1.361
H15-N14 0.949 0.997 0.992
H16-N14 0.950 0.998 0.995
Angle FIXDARa FIXDARb FIXCUK CHARMM HF/6-31G(d)
N2-C3-C5 123.5 121.3 122.4 120.7 122.2
C3-C5-C7 123.3 124.0 123.2 123.7 122.9
C5-C7-C9 108.1 108.6 109.5 109.6 110.5
C7-C9-C10 123.2 123.2 119.9 119.9 121.6
C9-C10-N2 121.7 121.7 123.4 123.1 123.4
C10-N2-C3 118.2 118.2 118.1 118.0 119.0
H1-N2-C3 120.3** 119.8 119.4
H4-C3-C5 118.8 118.8 122.6
H6-C5-C7 118.4 117.4 118.2
H8-C7-C9 115.6** 111.7 110.6
H17-C7-C9 105.6 107.8 111.1
H8-C7-H17 104.2** 108.8 105.7
H11-C10-C9 118.3 119.1 120.1
C7-C9-C12 122.6 122.7 120.1 122.2 122.3
C9-C12-013 119.7 120.4 120.3 122.1 123.2
C9-C12-N14 118.3 118.2 121.0 115.7 116.1
013-C12-N14 122.0 121.4 118.6 122.1 120.7
C12-N14-H15 120.0 121.8 121.9
C12-N14-H16 120.0 115.9 116.1
Dihedral Angle FIXDARa FIXDARb FIXCUK CHARMM HF/6-31G(d)
H1-N2-C3-H4 -0.5** 5.6 -12.5
N2-C3-C5-H6 173.6 -170.4 179.1
C3-C5-C7-H17 134.9** -112.6 117.9
H4-C3-C5-H6 -6.4 12.8 -0.1
H4-C3-C5-C7 173.6 169.1 -177.5
C5-C7-C9-C12 166.7 166.4 161.4 -148.9 -175.3
C5-C7-C9-C10 -12.9 -10.4 -18.9 7.8 4.4
C7-C9-C12-013 -163.9 -163.1 13.7 161.2 177.2
C7-C9-C12-N14 15.7 17.6 -162.5 -20.6 -1.6
C7-C9-C10-H11 -172.6 -175.4 -179.7
C7-C9-C10-N2 2.6 1.9 7.3 7.1 0.9
HS-C7-C9-C12 -72.7** -25.7 -53.9
H8-C7-C9-C10 107.0** 131.0 125.8
H17-C7-C9-C10 -139.4 109.5 -117.2
H17-C7-C9-C12 41.9 93.8 63.1
C9-C12-N14-H15 -0.1 1.6 -17.1
C9-C12-N14-H16 -180.0 178.7 -171.8
C9-C10-N2-H1 179.6** 174.5 -168.7
C9-C10-N2-C3 7.3 13.2 7.2 -26.1 -6.4
C10-N2-C3-C5 -4.8 -10.5 -8.0 29.3 6.0
C3-C5-C7-C9 15.5 13.1 18.7 4.6 -4.7
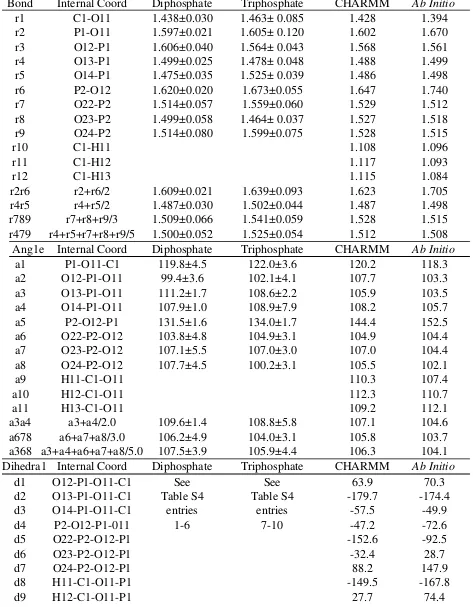
Table S3. Inorganic phosphate crystal, empirical and ab initio geometry data;bond lengths, angles and dihedral angles. Distances in and angles in degrees.
Bond Internal Coord Diphosphate Triphosphate CHARMM Ab Initio
r1 C1-O11 1.438±0.030 1.463± 0.085 1.428 1.394
r2 P1-O11 1.597±0.021 1.605± 0.120 1.602 1.670
r3 O12-P1 1.606±0.040 1.564± 0.043 1.568 1.561
r4 O13-P1 1.499±0.025 1.478± 0.048 1.488 1.499
r5 O14-P1 1.475±0.035 1.525± 0.039 1.486 1.498
r6 P2-O12 1.620±0.020 1.673±0.055 1.647 1.740
r7 O22-P2 1.514±0.057 1.559±0.060 1.529 1.512
r8 O23-P2 1.499±0.058 1.464± 0.037 1.527 1.518
r9 O24-P2 1.514±0.080 1.599±0.075 1.528 1.515
r10 C1-H11 1.108 1.096
r11 C1-H12 1.117 1.093
r12 C1-H13 1.115 1.084
r2r6 r2+r6/2 1.609±0.021 1.639±0.093 1.623 1.705
r4r5 r4+r5/2 1.487±0.030 1.502±0.044 1.487 1.498
r789 r7+r8+r9/3 1.509±0.066 1.541±0.059 1.528 1.515 r479 r4+r5+r7+r8+r9/5 1.500±0.052 1.525±0.054 1.512 1.508 Ang1e Internal Coord Diphosphate Triphosphate CHARMM Ab Initio
a1 P1-O11-C1 119.8±4.5 122.0±3.6 120.2 118.3
a2 O12-P1-O11 99.4±3.6 102.1±4.1 107.7 103.3
a3 O13-P1-O11 111.2±1.7 108.6±2.2 105.9 103.5
a4 O14-P1-O11 107.9±1.0 108.9±7.9 108.2 105.7
a5 P2-O12-P1 131.5±1.6 134.0±1.7 144.4 152.5
a6 O22-P2-O12 103.8±4.8 104.9±3.1 104.9 104.4
a7 O23-P2-O12 107.1±5.5 107.0±3.0 107.0 104.4
a8 O24-P2-O12 107.7±4.5 100.2±3.1 105.5 102.1
a9 H11-C1-O11 110.3 107.4
a10 H12-C1-O11 112.3 110.7
a11 H13-C1-O11 109.2 112.1
a3a4 a3+a4/2.0 109.6±1.4 108.8±5.8 107.1 104.6
a678 a6+a7+a8/3.0 106.2±4.9 104.0±3.1 105.8 103.7
a368 a3+a4+a6+a7+a8/5.0 107.5±3.9 105.9±4.4 106.3 104.1 Dihedra1 Internal Coord Diphosphate Triphosphate CHARMM Ab Initio
d1 O12-P1-O11-C1 See See 63.9 70.3
d2 O13-P1-O11-C1 Table S4 Table S4 -179.7 -174.4
d3 O14-P1-O11-C1 entries entries -57.5 -49.9
d4 P2-O12-P1-011 1-6 7-10 -47.2 -72.6
d5 O22-P2-O12-P1 -152.6 -92.5
d6 O23-P2-O12-P1 -32.4 28.7
d7 O24-P2-O12-P1 88.2 147.9
d8 H11-C1-O11-P1 -149.5 -167.8
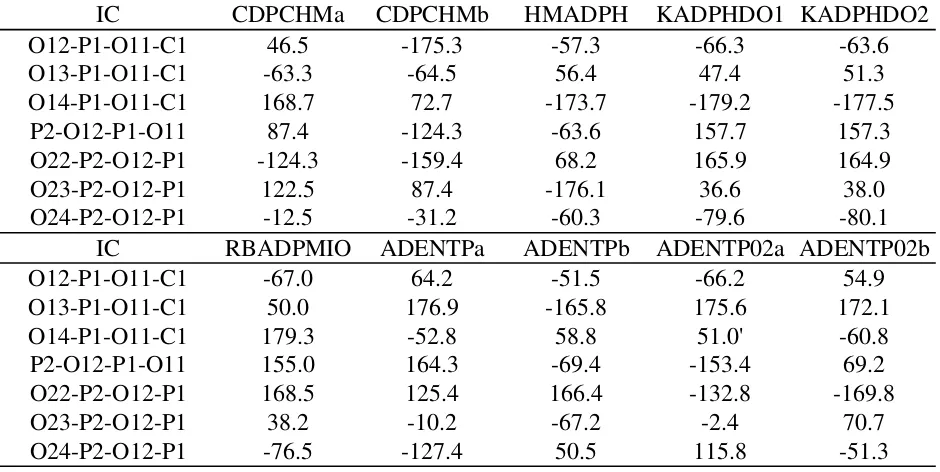
[image:6.612.88.561.104.711.2]Table S4 . Dihedral angles for the Diphosphate and Triphosphate crystal structures.
IC CDPCHMa CDPCHMb HMADPH KADPHDO1 KADPHDO2
O12-P1-O11-C1 46.5 -175.3 -57.3 -66.3 -63.6
O13-P1-O11-C1 -63.3 -64.5 56.4 47.4 51.3
O14-P1-O11-C1 168.7 72.7 -173.7 -179.2 -177.5
P2-O12-P1-O11 87.4 -124.3 -63.6 157.7 157.3
O22-P2-O12-P1 -124.3 -159.4 68.2 165.9 164.9
O23-P2-O12-P1 122.5 87.4 -176.1 36.6 38.0
O24-P2-O12-P1 -12.5 -31.2 -60.3 -79.6 -80.1
IC RBADPMIO ADENTPa ADENTPb ADENTP02a ADENTP02b
O12-P1-O11-C1 -67.0 64.2 -51.5 -66.2 54.9
O13-P1-O11-C1 50.0 176.9 -165.8 175.6 172.1
O14-P1-O11-C1 179.3 -52.8 58.8 51.0' -60.8
P2-O12-P1-O11 155.0 164.3 -69.4 -153.4 69.2
O22-P2-O12-P1 168.5 125.4 166.4 -132.8 -169.8
O23-P2-O12-P1 38.2 -10.2 -67.2 -2.4 70.7
O24-P2-O12-P1 -76.5 -127.4 50.5 115.8 -51.3
Cambridge crystal database identifiers: CDPCHM: Cytidine 5'-diphosphate choline monohydrate, HMADPH: tris(Hydroxymethyl)-methylammonium
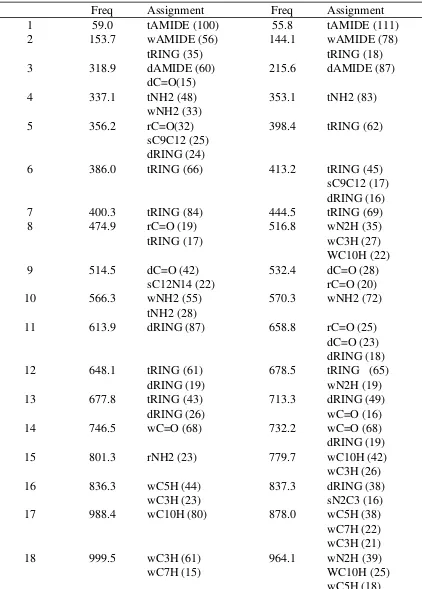
Table S5. Vibrational frequency assignments and relative contributions of the assignments to the frequency for NIC+. Frequencies in cm-1
Mode Ab initio CHARMM
Freq Assignment Freq Assignment
1 59.0 tAMIDE (100) 55.8 tAMIDE (111)
2 153.7 wAMIDE (56) 144.1 wAMIDE (78)
tRING (35) tRING (18)
3 318.9 dAMIDE (60) 215.6 dAMIDE (87)
dC=O(15)
4 337.1 tNH2 (48) 353.1 tNH2 (83)
wNH2 (33)
5 356.2 rC=O(32) 398.4 tRING (62)
sC9C12 (25) dRING (24)
6 386.0 tRING (66) 413.2 tRING (45)
sC9C12 (17) dRING (16)
7 400.3 tRING (84) 444.5 tRING (69)
8 474.9 rC=O (19) 516.8 wN2H (35)
tRING (17) wC3H (27)
WC10H (22)
9 514.5 dC=O (42) 532.4 dC=O (28)
sC12N14 (22) rC=O (20)
10 566.3 wNH2 (55) 570.3 wNH2 (72)
tNH2 (28)
11 613.9 dRING (87) 658.8 rC=O (25)
dC=O (23) dRING (18)
12 648.1 tRING (61) 678.5 tRING (65)
dRING (19) wN2H (19)
13 677.8 tRING (43) 713.3 dRING (49)
dRING (26) wC=O (16)
14 746.5 wC=O (68) 732.2 wC=O (68)
dRING (19)
15 801.3 rNH2 (23) 779.7 wC10H (42)
wC3H (26)
16 836.3 wC5H (44) 837.3 dRING (38)
wC3H (23) sN2C3 (16)
17 988.4 wC10H (80) 878.0 wC5H (38)
wC7H (22) wC3H (21)
18 999.5 wC3H (61) 964.1 wN2H (39)
wC7H (15) WC10H (25)
19 1008.4 dRING (32) 970.1 dC5H (23)
sC5C7 (23) sN2C3 (22)
sC3C5 (17)
20 1016.1 dRING (30) 1007.9 rNH2 (74)
sN2C3 (27)
21 1040.5 wC7H (60) 1024.1 dRING (47)
wC5H (37) sC5C7 (15)
22 1085.9 sC5C7(29) 1066.2 wC7H (42)
sC10N2 (28) wC5H (35)
23 1124.6 sC3C5 (18) 1073.3 sC10N2 (28)
sC9C10 (15) sC5C7 (23)
sC3C5 (20)
24 1142.9 sC3C5 (23) 1120.9 dC5H (42)
sC7C9 (21) dRING (24)
sN2C3 (16)
25 1200.2 dC5H (28) 1208.2 dN2H (25)
sC10N2 (22) sN2C3 (17)
26 1270.0 dN2H (32) 1273.7 dN2H (25)
dC3H (24) dC10H (18)
27 1317.8 dC10H (21) 1479.3 scNH2 (28)
dC7H (21) sC9C12 (25)
28 1323.4 rNH2 (23) 1554.5 dN2H (19)
dC7H (20) sN2C3 (18)
sC10N2 (16)
29 1429.7 sC12N14 (39) 1569.1 sC9C10 (33)
sC7C9 (25)
30 1468.8 dC3H (20) 1604.8 sC5C7 (20)
dC5H (20) sC3C5 (18)
31 1554.0 dN2H (27) 1665.0 scNH2 (55)
sC12N14 (19)
32 1613.1 sC7C9 (20) 1948.6 sC=O (64)
scNH2 (18) sC12N14 (21)
33 1647.2 sC3C5 (20) 1997.4 dC3H (36)
sC9C10 (17) dC10H (29)
dC7H (21)
34 1674.0 scNH2 (53) 2046.9 dC3H (49)
dN2H (19) dC10H (28)
35 1966.7 sC=O (71) 2080.7 dC7H (51)
wN2H (29) dC10H (26)
36 3067.2 sC7H (91) 2993.2 sC5H (64)
sC7H (21)
37 3084.6 sC5H (77) 2995.2 sC7H (77)
38 3100.2 sC3H (84) 2997.8 sC3H (90)
39 3102.7 sC10H (99) 2999.2 sC10H (70)
40 3420.0 sN2H (100) 3446.5 sNH2 (99)
41 3446.9 sNH2 (86) 3457.3 sN2H (99)
42 3583.2 sNH2a (85) 3562.3 1 sNH2a (99)
Stretching modes are represented by an "s" followed by the atoms in the bond stretch. For example, sC7H is the stretching niode of the hydrogens attached the C7 carbon. Angle deformations are represented by a "d" and a descriptor as in dRING, which are the ring angle deformations, and dN2H which are the amide hydrogen angle deformations. Wagging is represented by a "w" and a descriptor such as wAMIDE which is the amide carbon improper dihedral wag and wC7H which is the wagging of the hydrogens attached to atom C7. Other abbreviations: "t" for dihedral torsion, "r" for rocking, "sc" for
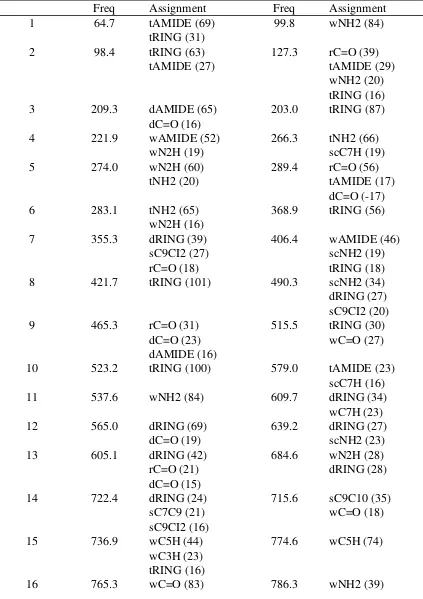
Table S6. Vibrational frequency assignments and relative contributions of the assignments to the frequency for NICH. Frequencies in cm-1
Mode Ab initio CHARMM
Freq Assignment Freq Assignment
1 64.7 tAMIDE (69) 99.8 wNH2 (84)
tRING (31)
2 98.4 tRING (63) 127.3 rC=O (39)
tAMIDE (27) tAMIDE (29)
wNH2 (20) tRING (16)
3 209.3 dAMIDE (65) 203.0 tRING (87)
dC=O (16)
4 221.9 wAMIDE (52) 266.3 tNH2 (66)
wN2H (19) scC7H (19)
5 274.0 wN2H (60) 289.4 rC=O (56)
tNH2 (20) tAMIDE (17)
dC=O (-17)
6 283.1 tNH2 (65) 368.9 tRING (56)
wN2H (16)
7 355.3 dRING (39) 406.4 wAMIDE (46)
sC9CI2 (27) scNH2 (19)
rC=O (18) tRING (18)
8 421.7 tRING (101) 490.3 scNH2 (34)
dRING (27) sC9CI2 (20)
9 465.3 rC=O (31) 515.5 tRING (30)
dC=O (23) wC=O (27)
dAMIDE (16)
10 523.2 tRING (100) 579.0 tAMIDE (23)
scC7H (16)
11 537.6 wNH2 (84) 609.7 dRING (34)
wC7H (23)
12 565.0 dRING (69) 639.2 dRING (27)
dC=O (19) scNH2 (23)
13 605.1 dRING (42) 684.6 wN2H (28)
rC=O (21) dRING (28)
dC=O (15)
14 722.4 dRING (24) 715.6 sC9C10 (35)
sC7C9 (21) wC=O (18)
sC9CI2 (16)
15 736.9 wC5H (44) 774.6 wC5H (74)
wC3H (23) tRING (16)
sC9C10 (34) wC=O (16)
17 902.1 sC5C7 (64) 835.7 sC5C7 (47)
18 958.9 dRING (33) 889.3 dRING (34)
sN2C3 (22) sC9C10 (31)
sC7C9 (19)
19 972.3 wC3H (58) 979.3 wC3H (24)
wC5H (28) sN2C3 (17)
20 977.9 wC10H (31) 995.9 wC10H (26)
wC3H (18) wC3H (26)
21 982.4 wC10H (55) 1004.7 rNH2 (49)
sNH2 (45) scNH2 (-16)
22 1011.4 tC7H (64) 1034.4 DRING (46)
sC9C12 (24) sC9C10 (-20)
23 1077.4 rNH2 (35) 1092.5 wC3H (23)
sC12N14 (35) wN2H (17)
WC10H (16)
24 1100.5 dRING (32) 1142.8 wAMIDE (41)
rNH2 (25) sC12N14 (29)
sC9C12 (16)
25 1150.0 sC10N2 (32) 1176.1 wN2H (34)
wC3H (27) wC10H (22)
26 1206.4 dC3H (26) 1218.0 dN2H (23)
dC5H (24) dC3H (21)
sC10N2 (16)
27 1215.1 wC7H (77) 1327.9 tC7H (50)
sC9C12 (34) dAMIDE (24) tRING (23) wC7H (-18)
28 1303.4 dC10H (40) 1388.7 sC9C12 (38)
rC7H (32) scNH2 (24)
tC7H (21) dC10H (16)
29 1349.3 sC12N14 (23) 1415.5 dC5H (37)
rC7H (22) tC7H (-18) dC3H (18)
30 1382.6 rC7H (28) 1476.6 tC7H (63)
sC7C9 (18) rC7H (-40)
dRING (19)
31 1402.3 dC3H (43) 1531.2 rC7H (41)
dC5H (28) wC7H (34)
32 1494.5 dN2H (57) 1551.2 rC7H (22)
sC10N2 (16) dRING (17)
33 1498.2 scC7H (97) 1558.3 dN2H (43)
34 1619.3 scNH2 (78) 1653.4 dC3H (31)
sN2C3 (26) sC3C5 (23)
35 1636.3 sC9C10 (46) 1667.7 rNH2 (52)
scNH2 (-18) sNH2a (17)
36 1717.5 sC3C5 (54) 1744.8 dC10H (28)
sC=O (24)
37 1749.8 sC=O (53) 1962.5 dC=O (109)
rC=O (-39)
38 2815.6 sC7Hsym (60) 2802.8 sC7C9 (53)
sC7Hasy (40) sC7Hsym (25)
sC7Hasy (24)
39 2850.2 sC7Hasy (60) 2843.9 sC7C9 (50)
sC7Hsym (40) sC7Hsym (27)
sC7Hasy (24)
40 3033.4 sC5H (86) 2989.6 sC5H (99)
41 3063.6 sC3H (86) 3092.2 sC10H (96)
42 3085.7 sC10H (99) 3092.7 sC3H (96)
43 3457.6 sNH2 (98) 3444.4 sNH2a (48)
sC7Hsym (22) sC7Hasy (22) scNH2 (18)
44 3520.6 sN2H (100) 3454.6 sN2H (99)
45 3572.5 sNH2a (98) 3566.2 sNH2a (38)
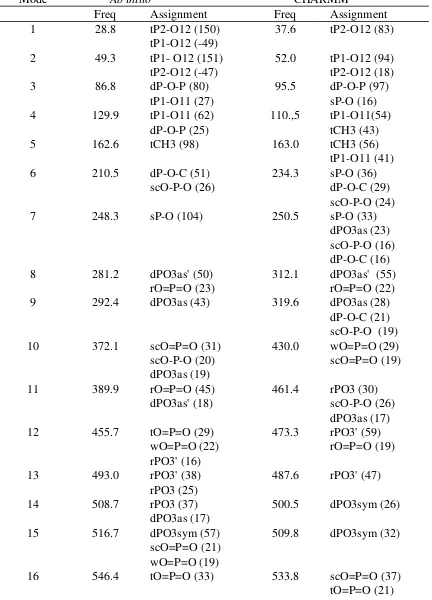
Table S7. Vibrational frequency assignments and relative contributions for methyl diphosphate. Frequencies in cm-1
Mode Ab initio CHARMM
Freq Assignment Freq Assignment
1 28.8 tP2-O12 (150) 37.6 tP2-O12 (83)
tP1-O12 (-49)
2 49.3 tP1- O12 (151) 52.0 tP1-O12 (94)
tP2-O12 (-47) tP2-O12 (18)
3 86.8 dP-O-P (80) 95.5 dP-O-P (97)
tP1-O11 (27) sP-O (16)
4 129.9 tP1-O11 (62) 110.,5 tP1-O11(54)
dP-O-P (25) tCH3 (43)
5 162.6 tCH3 (98) 163.0 tCH3 (56)
tP1-O11 (41)
6 210.5 dP-O-C (51) 234.3 sP-O (36)
scO-P-O (26) dP-O-C (29)
scO-P-O (24)
7 248.3 sP-O (104) 250.5 sP-O (33)
dPO3as (23) scO-P-O (16) dP-O-C (16)
8 281.2 dPO3as' (50) 312.1 dPO3as' (55)
rO=P=O (23) rO=P=O (22)
9 292.4 dPO3as (43) 319.6 dPO3as (28)
dP-O-C (21) scO-P-O (19)
10 372.1 scO=P=O (31) 430.0 wO=P=O (29)
scO-P-O (20) scO=P=O (19)
dPO3as (19)
11 389.9 rO=P=O (45) 461.4 rPO3 (30)
dPO3as' (18) scO-P-O (26)
dPO3as (17)
12 455.7 tO=P=O (29) 473.3 rPO3' (59)
wO=P=O (22) rO=P=O (19)
rPO3' (16)
13 493.0 rPO3' (38) 487.6 rPO3' (47)
rPO3 (25)
14 508.7 rPO3 (37) 500.5 dPO3sym (26)
dPO3as (17)
15 516.7 dPO3sym (57) 509.8 dPO3sym (32)
scO=P=O (21) wO=P=O (19)
16 546.4 tO=P=O (33) 533.8 scO=P=O (37)
17 586.6 dPO3sym (32) 635.6 scO=P=O (22)
sP-0 (20) sP-O (22)
scO=P=O (19)
18 684.4 sP-O (58) 730.4 sP-O (37)
wO=P=O (29) wO=P=O (28)
sP=O (20)
19 877.9 sP=O (65) 887.2 sP=O (65)
sP-O (35) sP-O (26)
20 957.9 sP=O (67) 1020.3 sP=O (45)
sP-O (30) sP-O (32)
21 1043.0 sP=O (65) 1054.1 sC-O (67)
sP-O (31)
22 1067.0 sP=O (92) 1057.9 dCH3as' (43)
dCH3as (31) rCH3' (17)
23 1080.9 sP=O (94) 1124.5 sP=O (70)
sP-O (30)
24 1101.1 sC-O (84) 1143.8 SP=O (91)
25 1145.7 sP=O (93) 1146.6 sP=O (90)
26 1170.0 dCH3as'(67) 1192.7 dCH3as (46)
dCH3as (29) dCH3a.s' (22)
rCH3' (15)
27 1204.7 dCH3as (61) 1256.9 sP=O (91)
dCH3as' (25)
28 1454.7 dCH3s (50) 1436.3 rCH3 (78)
rCH3' (42) dCH3as (17)
29 1488.6 dCH3s (49) 1470.9 rCH3' (61)
rCH3' (35) dCH3as' (28)
30 1506.6 rCH3 (82) 1638.4 dCH3s (84)
rCH3' (16)
31 2767.0 sC-H (100) 2853.7 sC-H (100)
32 2813.4 sC-H (100) 2913.9 sC-H (100)
33 2942.7 sC-H (100) 2916.1 sC-H (100)
Table S8. NIC+/NICH dipole moments in Debye.
ab initio Model 1 Model 2
NIC+ trans
Total 8.26 7.77 7.17
x -6.66 -6.09 -5.78
y -4.62 -4.76 -4.13
z 1.64 0.77 0.98
NIC+ cis
Total 6.98 6.17
x 6.57 6.11
y 1.77 0.84
z 1.56 -0.01
ab initio emp
NICH trans
total 4.82 3.65
x -2.50 -1.23
y -4.02 -3.40
z 0.94 -0.51
NICH cis
Total 3.72 2.18
x 3.21 1.10
y 1.83 1.88
Table S9. Unitcell parameters and energies of the NAD+-Li+ crystal minimizations for both Models 1 and 2. Energies in kcal/mol, distances in , angles in degrees and forces in
mdyn.
Cutoff a b c Total Energy Lattice Energy GRMS
VDWElec Model 1
exper 10-073 15-839 17.821
10-9-7 10.349 15.477 17.311 -554.94 -36.96 -215.61 0.00053 13-12-10 9.984 15.418 17-887 -595.25 -45.10 -202.82 0.04190 16-15-13 10.130 15.452 17.679 -614.18 -46.36 -201.01 0.00071 19-18-16 10.318 15.428 17.654 -624.51 -47.63 -198.40 0.00057 22-21-19 10.122 15.462 17.994 -629.21 -47.21 -196-06 0.00068 25-24-22 10.112 15.346 17.918 -633.75 -48.68 -192.96 0.00027 Model 2
exper 10.073 15.839 17.821
10-9-7 9.684 15.274 18.658 -542.84 -37.25 -226.71 0.00084 13-12-10 9.862 15.581 17.964 -587.27 -44.14 -214.97 0.00063 16-15-13 9.906 15.446 18.264 -604.96 -47.22 -209.56 0.00048 19-18-16 10.088 15.352 17.862 -614.57 -47.08 -204.73 0.00042 22-21-19 9.926 15.473 18.170 -620.91 -47.94 -202.44 0.00054 25-24-22 9.924. 15.352 18.188 -624.53 -47.83 -200.82 0.00056
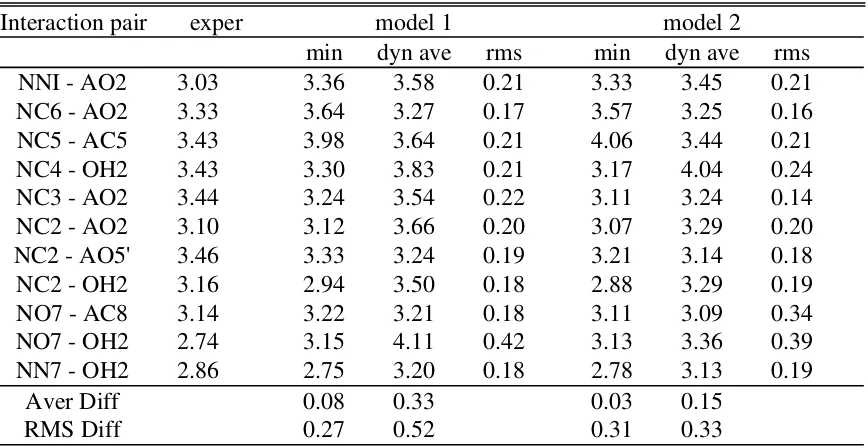
Table S10. Interaction distances involving the nicotinamide ring in the NAD-LI+ crystal
from x-ray crystallography and the calculations using Models 1 and 2.
Interaction pair exper model 1 model 2
min dyn ave rms min dyn ave rms
NNI - AO2 3.03 3.36 3.58 0.21 3.33 3.45 0.21
NC6 - AO2 3.33 3.64 3.27 0.17 3.57 3.25 0.16
NC5 - AC5 3.43 3.98 3.64 0.21 4.06 3.44 0.21
NC4 - OH2 3.43 3.30 3.83 0.21 3.17 4.04 0.24
NC3 - AO2 3.44 3.24 3.54 0.22 3.11 3.24 0.14
NC2 - AO2 3.10 3.12 3.66 0.20 3.07 3.29 0.20
NC2 - AO5' 3.46 3.33 3.24 0.19 3.21 3.14 0.18
NC2 - OH2 3.16 2.94 3.50 0.18 2.88 3.29 0.19
NO7 - AC8 3.14 3.22 3.21 0.18 3.11 3.09 0.34
NO7 - OH2 2.74 3.15 4.11 0.42 3.13 3.36 0.39
NN7 - OH2 2.86 2.75 3.20 0.18 2.78 3.13 0.19
Aver Diff 0.08 0.33 0.03 0.15
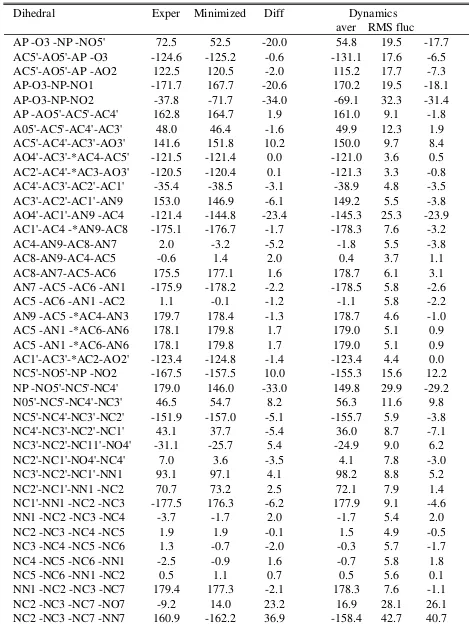
[image:18.612.90.524.480.704.2]Table S11. NAD crystal experimental and calculated dihedrals for Model 2 with the final diphosphate parameters. Angles indegrees.
Dihedral Exper Minimized Diff Dynamics
aver RMS fluc
AP -O3 -NP -NO5' 72.5 52.5 -20.0 54.8 19.5 -17.7
AC5'-AO5'-AP -O3 -124.6 -125.2 -0.6 -131.1 17.6 -6.5
AC5'-AO5'-AP -AO2 122.5 120.5 -2.0 115.2 17.7 -7.3
AP-O3-NP-NO1 -171.7 167.7 -20.6 170.2 19.5 -18.1
AP-O3-NP-NO2 -37.8 -71.7 -34.0 -69.1 32.3 -31.4
AP -AO5'-AC5'-AC4' 162.8 164.7 1.9 161.0 9.1 -1.8
A05'-AC5'-AC4'-AC3' 48.0 46.4 -1.6 49.9 12.3 1.9
AC5'-AC4'-AC3'-AO3' 141.6 151.8 10.2 150.0 9.7 8.4
AO4'-AC3'-*AC4-AC5' -121.5 -121.4 0.0 -121.0 3.6 0.5 AC2'-AC4'-*AC3-AO3' -120.5 -120.4 0.1 -121.3 3.3 -0.8 AC4'-AC3'-AC2'-AC1' -35.4 -38.5 -3.1 -38.9 4.8 -3.5
AC3'-AC2'-AC1'-AN9 153.0 146.9 -6.1 149.2 5.5 -3.8
AO4'-AC1'-AN9 -AC4 -121.4 -144.8 -23.4 -145.3 25.3 -23.9 AC1'-AC4 -*AN9-AC8 -175.1 -176.7 -1.7 -178.3 7.6 -3.2
AC4-AN9-AC8-AN7 2.0 -3.2 -5.2 -1.8 5.5 -3.8
AC8-AN9-AC4-AC5 -0.6 1.4 2.0 0.4 3.7 1.1
AC8-AN7-AC5-AC6 175.5 177.1 1.6 178.7 6.1 3.1
AN7 -AC5 -AC6 -AN1 -175.9 -178.2 -2.2 -178.5 5.8 -2.6
AC5 -AC6 -AN1 -AC2 1.1 -0.1 -1.2 -1.1 5.8 -2.2
AN9 -AC5 -*AC4-AN3 179.7 178.4 -1.3 178.7 4.6 -1.0
AC5 -AN1 -*AC6-AN6 178.1 179.8 1.7 179.0 5.1 0.9
AC5 -AN1 -*AC6-AN6 178.1 179.8 1.7 179.0 5.1 0.9
AC1'-AC3'-*AC2-AO2' -123.4 -124.8 -1.4 -123.4 4.4 0.0 NC5'-NO5'-NP -NO2 -167.5 -157.5 10.0 -155.3 15.6 12.2 NP -NO5'-NC5'-NC4' 179.0 146.0 -33.0 149.8 29.9 -29.2
N05'-NC5'-NC4'-NC3' 46.5 54.7 8.2 56.3 11.6 9.8
NC5'-NC4'-NC3'-NC2' -151.9 -157.0 -5.1 -155.7 5.9 -3.8
NC4'-NC3'-NC2'-NC1' 43.1 37.7 -5.4 36.0 8.7 -7.1
NC3'-NC2'-NC11'-NO4' -31.1 -25.7 5.4 -24.9 9.0 6.2
NC2'-NC1'-NO4'-NC4' 7.0 3.6 -3.5 4.1 7.8 -3.0
NC3'-NC2'-NC1'-NN1 93.1 97.1 4.1 98.2 8.8 5.2
NC2'-NC1'-NN1 -NC2 70.7 73.2 2.5 72.1 7.9 1.4
NC1'-NN1 -NC2 -NC3 -177.5 176.3 -6.2 177.9 9.1 -4.6
NN1 -NC2 -NC3 -NC4 -3.7 -1.7 2.0 -1.7 5.4 2.0
NC2 -NC3 -NC4 -NC5 1.9 1.9 -0.1 1.5 4.9 -0.5
NC3 -NC4 -NC5 -NC6 1.3 -0.7 -2.0 -0.3 5.7 -1.7
NC4 -NC5 -NC6 -NN1 -2.5 -0.9 1.6 -0.7 5.8 1.8
NC5 -NC6 -NN1 -NC2 0.5 1.1 0.7 0.5 5.6 0.1
NN1 -NC2 -NC3 -NC7 179.4 177.3 -2.1 178.3 7.6 -1.1
NC2 -NC3 -NC7 -NO7 -9.2 14.0 23.2 16.9 28.1 26.1
NP -O3 -AP -AO5' 132.5 164.6 32.1 161.1 29.6 28.6
O3 -NP -NO5'-NC5' 79.1 78.7 -0.4 81.3 9.9 2.2
RMS difference 12.8 12.7
Table S12. NAD crystal experimental and calculated interaction distances for Model 2. Distances given in .
Interaction pair exper Minimized Diff Dynamics Diff Aver rms flu
NAD1 NC6 -NAD1 N05' 3.23 3.52 0.30 3.54 0.23 0.31
NAD1 AN7 -NAD1 NO1 3.22 3.73 0.52 3.76 0.14 01.54
NAD1 AC8 -NAD1 O3 3.45 3.23 -0.21 3.35 0.13 -0.10
NAD1 AC8 -NAD1 NO1 3.41 4.07 0.65 4.02 0.17 0.60
NAD1 AO1 -NAD1 NO2 2.95 2.76 -0.19 2.76 0.11 -0.19
NAD1 AO5'-NAD1 AC2' 3.22 3.06 -0.16 3.13 0.10 -0.08
NAD1 AO2'-WATX OH2 2.66 2.71 0.05 2.78 0.12 0.12
NAD1 NN1 -C009 AO2 3.03 3.35 0.32 3.28 0.15 0.25
NAD1 NC6 -C009 AO2 3.33 3.59 0.26 3.49 0.23 0.16
NAD1 NC5 -C020 AC5 3.43 4.07 0.65 4.08 0.22 0.65
NAD1 NC4 -C020 OH2 3.43 3.15 -0.28 3.21 0.13 -0.22
NAD1 NC3 -C009 AO2 3.44 3.10 -0.35 3.27 0.25 -0.17
NAD1 NC2 -C009 AO2 3.10 3.07 -0-03 3.14 0.19 0.03
NAD1 NC2 -C009 AO5' 3.46 3.23 -0.24 3.25 0.16 -0.21
NAD1 NC2 -C020 OH2 3.16 2.89 -0.27 2.98 0.19 -0.18
NAD1 AN7 -C008 OH2 3.38 3.30 -0.08 3.27 0.28 -0.11
NAD1 AN9 -C020 NO4' 3.19 3.00 -0.20 3.09 0.15 -0.10
NAD1 AN1 -C003 NC2' 3.44 3.73 0.29 3.87 0.22 0.42
NAD1 AN1 -C003 NO2' 2.77 2.93 0.15 3.04 0.16 0.26
NAD1 AC2 -C003 NO3' 3.38 3.12 -0.25 3.28 0.15 -0.10
NAD1 AN3 -C020 NO4' 3.16 3.87 0.71 3.71 0.18 0.54
NAD1 AC4 -C020 NO4' 3.15 3.31 0.16 3.32 0.16 0.17
NAD1 AN6 -C003 NO2Õ 3.41 3.19 -0.22 3.40 0.17 -0.01
NAD1 AN6 -C008 AO2' 3.09 3.02 -0.07 3.12 0.15 0.03
NAD1 AN6 -C008 AO3' 2.91 2.95 0.04 3.09 0.22 0.18
NAD1 AO1 -C020 AN7 3.14 3.16 0.01 3.06 0.10 -0.08
NAD1 AO1 -C020 AN6 2.89 2.73 -0.15 2.79 0.12 -0.10
NAD1 AO1 -C020 NO1 3.25 3.41 0.15 3.46 0.19 0.20
NAD1 NO2 -C020AN7 3.38 3.71 0.33 3.85 0.21 0.46
NAD1 NO2 -C020 NO1 3.13 3.62 0.49 3.67 0.13 0.53
NAD1 AO4'-C020 NO4' 2.94 3.08 0.14 3.17 0.16 0.23
NAD1 AO4'-C020 NC6 3.43 2.99 -0.43 3.21 0.18 -0.22
NAD1 AC1'-C015 AO2 3.31 3.35 0.04 3.44 0.13 0.13
NAD1 AC1'-C020 NO4' 3.16 3.31 0.15 3.27 0.12 0.12
NAD1 AO3'-C015 AO2 2.59 2.65 0.06 2.71 0.10 0.12
NAD1 AO3'-C015 NC3' 3.24 3.20 -0.04 3.22 0.10 -0.02
NAD1 AO3'-C015 NO3' 2.70 2.68 -0.01 2.70 0.08 0.00
NAD1 AC5'-C020 NO1 3.48 3.52 0.04 3.64 0.25 0.16
Interactions involving the amide group
NAD1 NO7 -C020 OH2 2.74 3.12 0.38 3.20 0.26 0.46
NAD1 NN7 -C020 OH2 2.86 2.79 -0.07 2.95 0.35 0.09
NAD1 NO2 -C013 NN7 2.88 2.85 -0.04 2.93 0.17 0.05
Interactions involving the Lithium ion
ION1 LIT -NAD1 AC5 3.19 3.12 -0.08 3.25 0.13 0.06
ION1 LIT -NAD1 AN7 2.13 2.28 0.15 2.37 0.15 0.24
ION1 LIT -NAD1 ACS 3.01 3.08 0.07 3.12 0.15 0.11
ION1 LIT -NAD1 NP 3.31 3.21 -0-10 3.27 0.08 -0.04
ION1 LIT -NAD1 NO1 1.92 2.05 0.12 2.07 0.06 0.15
ION1 LIT -NAD1 AH8 3.17 3.47 0.30 3.44 0.18 0.27
ION1 L1T -NAD1 AH61 2.75 2.84 0.08 2.98 0.17 0.23
NAD1 AO1 -C020 LIT 1.88 2.13 0.24 2.14 0.08 0.26
NAD1 NO2 -C020 LIT 1.86 2.08 0.23 2.12 0.07 0.26
Average difference 0.07 0.16
RMS difference 0.27 0.26
Table S13) Alcohol Dehydrogenase bound NAD experimental and calculated dihedrals. Dihedral angles in degrees.
Dihedral Experimental Dynamics Difference
Aver RMS flu
AP -O3 -NP -NO5' -155.7 -173.8 11.9 -18.1
AC5'-AO5'-AP -O3 -85.4 53.8 10.8 139.2
AC5'-AO5'-AP -AO2 36.5 -178.7 10.8 144.8
AP -O3 -NP -NO1 96.4 75.6 12.4 -20.8
AP -O3 -NP -NO2 -36.5 -61.9 12.6 -25.4
AP -AO5'-AC5'-AC4' -137.7 166.5 14.2 -55.8
AO1 -AP -AO5'-AC5' 163.4 -55.9 11.3 140.8
AO5'-AC5'-AC4'-AC3' 47.5 -64.6 7.8 -112.1
AC5'-AC4'-AC3'-AO3' 103.6 124.8 8.1 21.2
AO4'-AC3'-*AC4-AC5' -11.5 18.4 14.8 29.9
AC4'-AC3'-AC2'-AC1' -17.5 -27.1 5.4 -9.6
AC3'-AC2'-AC1'-AN9 167.5 157.9 10.9 -9.6
AO4'-AC1'-AN9 -AC4 -107.7 -124.8 17.1 -17.1
AC1'-AC4 -*AN9-AC8 -174.4 175.1 8.9 -10.5
AC4 -AN9 -AC8 -AN7 0.6 0.0 4.7 -0.6
AC8 -AN9 -AC4 -AC5 -0.9 0.2 3.9 1.1
AC8 -AN7 -AC5 -AC6 178.1 -180.0 5.4 1.9
AN7 -AC5 -AC6 -AN1 -178.4 -178.5 6.4 -0.1
AC5 -AC6 -AN1 -AC2 -0.5 -1.4 5.6 -0.9
AN9 -AC5 -*AC4-AN3 178.3 -179.9 5.0 1.8
AC5 -AN1 -*AC6-AN6 179.2 179.3 5.2 0.1
AC5 -AN1 -*AC6-AN6 179.2 179.3 5.2 0.1
AC1'-AC3'-*AC2-AO2' 96.0 126.9 39.6 30.9
NC5'-NO5'-NP -NO2 -55.7 -54.8 9.1 0.9
NP -NO5'-NC5'-NC4' -170.9 -166.1 9.1 4.8
NO5'-NC5'-NC4'-NC3' 52.6 44.7 9.3 -7.9
NC5'-NC4'-NC3'-NC2' -95.0 -93.0 5.0 2.0
NC4'-NC3'-NC2'-NC1' -38.7 -38.1 3.8 0.6
NC3'-NC2'-NC1'-NO4' 45.4 37.5 5.1 -7.9
NC2'-NC1'-NO4'-NC4' -34.8 -22.0 7.0 12.8
NO2'-NC2'-NC1'-NO4' 167.3 160.3 5.1 -7.0
NC3'-NC2'-NC1'-NN1 163.2 156.4 5.1 -6.8
NO3'-NC3'-NC2'-NC1' 83.6 82.0 5.0 -1.6
NC2'-NC1'-NN1 -NC2 140.0 142.1 7.5 2.1
NC1'-NN1 -NC2 -NC3 173.3 166.9 7.6 -6.4
NN1 -NC2 -NC3 -NC4 1.0 0.2 5.7 -0.8
NC2 -NC3 -NC4 -NC5 -0.4 -2.0 5.3 -1.6
NC3 -NC4 -NC5 -NC6 -0.1 2.2 5.2 2.3
NC4 -NC5 -NC6 -NN1 0.0 -0.4 5.4 -0.4
NC5 -NC6 -NN1 -NC2 0.6 -1.4 5.8 -2.0
[image:24.612.83.562.103.721.2]NC2 -NC3 -NC7 -NO7 -160.6 -161.3 10.0 -0.7
NC2 -NC3 -NC7 -NN7 19.9 23.6 10.8 3.7
RMS Difference 43.3(14.2)
Table S14) Alcohol Dehydrogenase bound NAD experimental and calculated interaction distances. Distances in .
Interaction Experiment Dynamics Difference
Average RMS flu
AO4'- CG1 3.29 3.99 0.25 0.70
AC1'- OD1 3.41 3.86 0.29 0.45
AN3 - OD1 3.45 4.02 0.43 0.57
AN6 - NH1 3.03 3.63 0.49 0.60
AO2'- OD1 2.81 2.74 0.16 -0.07
AO3'- CG 3.45 3.56 0.17 0.11
AO3'- OD1 3.46 3.84 0.26 0.38
AO3'- OD2 2.68 2.63 0.10 -0.05
AO3'- NZ 2.77 2.85 0.14 0.08
AC5'- N 3.40 4.64 0.19 1.24
AO1 - NE 2.64 2.73 0.10 0.09
AO1 - CZ 3.23 3.16 0.08 -0.07
AO1 - NH2 3.00 2.71 0.11 -0.29
AO2 - CA 3.17 3.45 0.18 0.28
O3 - CB 3.28 3.38 0.22 0.10
NO1 - N 3.30 3.15 0.25 -0.15
NO1 - CB 3.36 3.61 0.25 0.25
NO1 - NH1 2.83 2.72 0.11 -0.11
NO2 - N 3.22 3.23 0.18 0.01
NO2 - N 2.95 2.95 0.15 0.00
NC5' - O 3.22 3.31 0.19 0.09
NO2' - OG 2.80 2.78 0.10 -0.02
NO2' - NE2 3.30 3.13 0.19 -0.17
NO2' - CD2 3.26 3.09 0.17 -0.17
NC3' - O 3.15 3.46 0.17 0.31
NO3' - NE2 3.15 3.19 0.24 0.04
NO3' - C 3.38 3.51 0.14 0.13
NO3' - O 2.57 2.74 0.13 0.17
NO3' - N 3.09 3.29 0.24 0.20
NC4' - O 3.39 3.68 0.23 0.29
NC5 - SG 3.35 3.37 0.12 0.02
NC4 - OG1 3.44 3.44 0.21 0.00
NC4 - CG2 3.44 3.40 0.15 -0.04
NC2 - O 2.93 3.01 0.14 0.08
NC5 - O1 3.09 3.15 0.18 0.06
NC4 - C1 3.36 3.60 0.24 0.24
NC4 - O1 3.31 3.33 0.18 0.02
Interactions involving the amide group
NC7 - CG2 3.48 3.89 0.22 0.41
[image:26.612.94.560.102.711.2]NN7 - O 2.98 2.96 0.15 -0.02
NN7 - O 2.93 3.03 0.20 0.10
Interactions involving the zinc ion
NC5 - ZN 3.34 3.50 0.14 0.16
Interactions involving water
AN1 - OH2 3.26 5.16 2.03 1.90
AN6 - OH2 3.12 7.05 3.02 3.93
AO2'- OH2 2.94 3.53 0.68 0.59
AO3'- OH2 3.20 3.96 0.85 0.76
AC5'- OH2 3.26 3.75 0.25 0.49
AO2 - OH2 2.72 2.67 0.11 -0.05
AO2 - OH2 2.65 2.80 0.17 0.15
NO2 - OH2 2.71 2.73 0.20 0.02
AO4'- OH2 3.09 7.20 4.86 4.11
AN7 - OH2 2.92 8.95 4.27 6.03
AC8 - OH2 3.40 6.26 1.47 2.86
AC2'- OH2 3.47 18.93 8.31 15.46
AO2'- OH2 2.65 19.56 8.57 16.91
AC5'- OH2 3.39 7.46 4.89 4.07
AO5'- OH2 2.73 8.42 4.59 5.69
Average Difference 1.21 (0.20)
RMS Difference 3.40 (0.28)
Table S15. Bond Parameters. Equilibrium bond lengths in and force constants are given in kcal/mol/.
bond type Kb bo
NIC+/NICH
CN1A CN3 302.0 1.480
CN1A NN1 560.0 1.360
CN1A ON1 860.0 1.230
CN3A CN3 450.0 1.360
CN3B CN3 420.0 1.350
CN3B NN2 420.0 1.315
CN3A HN3 350.0 1.09
CN3B HN3 350.0 1.09
CN3A HN3B 350.0 1.09
CN3B HN3B 350.0 1.09
NICH
CN3 CN8 222.5 1.490
CN3C NN2 420.0 1.355
CN3C HN3 374.0 1.09
CN3C CN3 420.0 1.320
Phosphate
ON2 P2 300.0 1.68
Table S16. Angle Parameters. Equilibrium bond angles are given in degrees and force constants are given in kcal/mol/.
angle type K KUB So
NIC+/NICH
CN3A CN3 CN3B 40.0 118.0
CN3A CN3 CN1A 40.0 110.2
CN3B CN3 CN1A 10.0 131.8
CN3 CN1A NN1 85.0 113.0 80.0 2.46
ON1 CN1A CN3 85.0 118.5 20.0 2.43
ON1 CNLA NN1 85.0 128.5 20.0 2.17
CN3B CN3 HN3 30.0 122.0
CN3A CN3 HN3 30.0 119.0
CN3 CN3A CN3 50.0 118.0
CN3 CN3A HN3 30.0 121.0
CN3 CN3B NN2 120.0 122.0
HN3 CN3B NN2 30.0 117.5
CN3 CN3B HN3 30.0 120.5
CN6 CN9 ON2 75.7 110.1
CN1A NN1 HN1 35.0 120.0
CN3B NN2 CN3B 30.0 120.0
CN3B NN2 CN6B 70.0 121.7
HN3B CN3B NN2 80.0 117.5
CN3 CN3A HN3B 80.0 121.0
CN3 CN3B HN3B 80.0 120.5
CN3B CN3 HN3B 30.0 122.0
CN3A CN3 HN3B 30.0 119.0
NICH
CN3 CN8 CN3 125.0 108.0
CN3 CN3 CNS 53.5 108.5
CN8 CN3 CN1A 125.0 124.2
CN3 CNS HN7 55.0 110.1
CN8 CN3 HN3 30.0 116.0
CN8 CN3 HN3B 30.0 122.0
CN3 CN3C NN2 60.0 122.0
CN3C NN2 CN3C 20.0 114.0
CN3C CN3 CN8 43.5 128.0
CN3 CN3C HN3 42.0 119.0
HN3 CN3C NN2 42.0 119.0
CN3C CN3 HN3 42.0 116.0
CN3C NN2 HN2 39.0 123.0
CN3C CN3 CN1A 5.0 107.8
CN3B CN3 CN8 53.5 108.5
CN3B NN2 HN2 32.0 117.4
Phosphate
P2 ON2 P 15.0 140.0 -40.0 2.800
P2 ON2 P2 15.0 140.0 -40.0 2.800
CN9 ON2 P2 20.0 120.0 35.0 2.33
ON2 P2 ON2 80.0 104.3
ON2 P2 ON3 98.9 111.6
Table S17. Dihedral Angle Parameters. Equilibrium dihedral angles in degrees force constants are given in kcal/mol/radian.
Dihedral K n
NIC+/NICH
CN3 NN2 CN3B HN3B 7.0 2 180.0
HN2 NN2 CN3B HN3B 3.0 2 180.0
HN3B CN3 CN3A HN3B 2.0 2 180.0
HN3B CN3 CN3B HN3B 1.0 2 180.0
NN2 CN3B CN3 HN3B 7.0 2 180.0
CN3 CN1A CN3 HN3B 7.0 2 180.0
CN1A CN3 CN3B HN3B 1.0 2 180.0
CN1A CN3 CN3A HN3B 5.0 2 180.0
CN3 CN1A CN3 HN3 7.0 2 180.0
CN1A CN3 CN3 HN3 1.0 2 180.0
CN3A CN3 CN1A ON1 2.38 2 180.0
CN3B CN3 CN1A ON1 2.38 2 180.0
CN3A CN3 CN1A NN1 0.35 1 180.0
CN3A CN3 CN1A NN1 0.62 2 0.0
CN3B CN3 CN1A NN1 0.35 1 0.0
CN3B CN3 CN1A NN1 0.62 2 0.0
CN3 CN3A CN3 CN1A 3.0 2 180.0
CN3 CN3A CN3 CN3B 6.0 2 180.0
NN2 CN3B CN3 CN3A 7.0 2 180.0
CN3B NN2 CN3B CN3 4.0 2 180.0
X CN3 CN3A X 1.0 2 180.0
X CN3 CN3B X 1.0 2 180.0
X CN3 CN8 X 1.0 3 180.0
X NN1 CN1A X 2.5 2 180.0
X NN2 CN3B X 1.0 2 180.0
NICH
CN8 CN3 CN1A ON1 1.00 2 180.0
CN8 CN3 CN1A ON1 1.00 3 0.0
C,N8 CN3 CN1A ON1 0.40 6 0.0
CN8 CN3 CN1A NN1 0.50 2 180.0
CN8 CN3 CN1A NN1 0.35 3 180.0
CN8 CN3 CN1A NN1 0.40 6 0.0
CN3C CN3 CN1A ON1 0.30 1 0.0
CN3C CN3 CN1A ON1 1.95 2 180.0
CN3C CN3 CN1A NN1 1.10 1 180.0
CN3C CN3 CN1A NN1 1.95 2 180.0
HN2 NN2 CN3C HN3 4.0 2 180.0
CN3 NN2 CN3C HN3 7.0 2 180.0
CN8 CN3 CN3C NN2 0.1 2 180.0
CN3C CN3 CN8 CN3 4.0 3 180.0
CN1A CN3 CN3C NN2 2.5 2 180.0
X CN3 CN3C X 0.1 2 180.0
X NN2 CN3C X 0.1 2 180.0
CN3C NN2 CN3C CN3 0.1 2 180.0
Phosphate
P2 ON2 P ON2 0.03 2 0.0
P2 ON2 P ON2 0.03 3 0.0
P ON2 P2 ON2 0.03 2 0.0
P ON2 P2 ON2 0.03 3 0.0
P2 ON2 P2 ON2 0.03 2 0.0
P2 ON2 P2 ON2 0.03 3 0.0
P ON2 P ON2 0.03 2 0.0
P ON2 P ON2 0.03 3 0.0
P ON2 P ON3 0.10 2 0.0
P ON2 P ON3 0.03 3 0.0
P ON2 P2 ON3 0.10 2 0.0
P ON2 P2 ON3 0.03 3 0.0
P2 ON2 P ON3 0.10 2 0.0
P2 ON2 P ON3 0.03 3 0.0
P2 ON2 P2 ON3 0.10 2 0.0
Table S18. Improper Dihedral Angle Parameters Equilibrium. Improper dihedral angles in degrees and force constant in are given in kcal/mol - A.
Improper Dihedral K o
NIC+/NICH
HN3B X X CN3 15.0 0.0
HN3B X X CN3A 13.0 0.0
HN3B X X CN3B 13.0 0.0
HN2 CN3 CN3B NN2 50.0 0.0
HN1 HN1 CNIA NN1 -5.0 0.0
ON1 X X CNIA 40.0 0.0
HN7 CN3 CN3 CN8 18.0 0.0
NICH
HN3 X X CN3C 53.0 0.0
Table S19. Nonbonded Parameters. Epsilon is in kcal/mol and Rmin are in
Atom Rmin/2
NIC+/NICH
CN3A -0.180 1.80
CN3B -0.180 1.80
CN1A -0.070 2.00
HN3B -0.046 0.90
HN3N -0.046 0.90
NICH
CN3C -0.180 1.80
phosphate
P2 -0.585 2.15
[image:33.612.91.327.325.492.2]Supplemental Figures
Figure S1. Adiabatic energy surface for the amide rotation in NIC+ for the empirical (F), HF/6-31G(d) (B) and MP2/6-31G(d)//6-31G(d) (E) levels of theory. Energies in
kcal/mol.
Figure S2. Adiabatic energy surface for the amide rotation in NICH for the empirical (F), HF/6-31G(d) (B) and MP2/6-31G(d)//6-31G(d) (E) levels of theory. Energies in
kcal/mol.
Figure S3. Angle variations versus amide dihedral rotation angle for NIC+ at the
empirical and HF/6-31G(d) levels of theory. Data is included for the C10-C9-C12 (emp. (J), HF/6-31G(d), (E)), C7-C9-C12 (emp. (B), HF/6-31G(d), (G)), and C9-C12-N14 (emp. (F), HF/6-31G(d), (A)) angles.
Figure S4. Angle variations versus amide dihedral rotation angle for NICH at the
Figure S1
B B
B B B
B B
B B
B
B
B
B
B
B
B B B B J
J
J
J
J
J J H
H
H
H
H
H H
0 1 2 3 4 5 6
0 30 60 90 120 150 180
Figure S2 F F F F F F F F F F F F F F F F F F F B B B B B B B B BB E E E E E E E E EE 0 1 2 3 4 5 6 7 8 9
0 30 60 90 120 150 180
Figure S3
J J J
J J
J J J J J
J J J J J J J J J
B B B B B B B B B B
B B B B B B B B B
F F F
F F
F F F F F F F
F F
F F
F F F E E E E E E E Ñ Ñ Ñ Ñ Ñ Ñ Ñ A A A A A A A 110 115 120 125 130
0 30 60 90 120 150 180
Figure S4
J J J
J J J
J J
J J
J J
J J
J J
J J J
B B B B B B B B B B B B B B B B B B B F F F F F F F F
F F F
F F F F F
F F F E E E E E E
E E EE
Ñ Ñ Ñ Ñ Ñ Ñ Ñ Ñ ÑÑ A A A A A A A A AA 114 116 118 120 122 124
0 30 60 90 120 150 180