Molecular and functional characterization of a rose lipoxygenase
cDNA related to flower senescence
Masako Fukuchi-Mizutani
a,*, Kanako Ishiguro
b, Toru Nakayama
b,c,
Yukiko Utsunomiya
b, Yoshikazu Tanaka
a, Takaaki Kusumi
a, Takashi Ueda
baInstitute for Fundamental Research,Suntory Ltd.,1-1-1Wakayamadai,Shimamoto-cho,Mishima-gun,Osaka,618-8503Japan bFaculty of Nutrition,Kobe Gakuin Uni6ersity,518Arise,Ikawadani-cho,Nishi-ku,Kobe,Hyogo,651-21Japan
cDepartment of Biochemistry and Engineering,Tohoku Uni6ersity,07Aoba Aza Aramaki,Aoba-ku,Sendai,Miyagi980-8579Japan
Received 20 June 2000; received in revised form 28 August 2000; accepted 29 August 2000
Abstract
A cDNA encoding lipoxygenase, Rlox1, was isolated from a cDNA library of senescing rose petals using tomato lipoxygenase cDNA fragments as probes. Characterization of the Rlox1 protein expressed inEcherichia colirevealed that the Rlox1 protein was a soluble lipoxygenase with an unusual optimal pH in the acidic region (pH 4.5 – 5.0). Northern blot analysis showed that the transcript of theRlox1 gene was dramatically increased in response to senescence of rose petals. Treatment of rose flowers with ethylene also elevated the mRNA of theRlox1 gene. These results suggest that the Rlox1 lipoxygenase is involved in senescence of rose flowers. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Rosa hybrida; Flower senescence; Lipoxygenase; Ethylene
www.elsevier.com/locate/plantsci
1. Introduction
Lipoxygenase (EC 1. 13.11.12) occurs widely in higher plants and catalyzes the dioxygenation of polyunsaturated fatty acids containing one or more cis, cis-1,4-pentadiene structure(s) [1,2]. The primary products of lipoxygenase-catalyzed reac-tions, fatty acid hydroxyperoxides, are further me-tabolized into molecules functioning in many biological processes, such as plant growth regula-tors, signal transduction molecules (e.g., jas-monates), and compounds responsible for green odor (hexanals) [3 – 5].
It has been reported that the expression of lipoxygenase genes is induced by stresses such as
insect feeding [6], pathogen infection [7 – 12], low water potential [13] and mechanical stress [6,14 – 18]. In addition, lipoxygenases are suggested to be involved in fruit ripening [19,20] and senescence of vegetative or reproductive organs [21,22].
Many physiological and biochemical changes, including ethylene production and degradation of cellular membranes, occur during flower senes-cence [23,24]. Rose is the most important cut flower and it is critical to control senescence of rose flowers during postharvest. Although several mechanistic studies on senescence of rose flowers have been reported [25 – 27], the molecular biology of this process remains to be clarified. We started a program to study the molecular biology of de-velopment and senescence of rose flowers, and found that a novel gene encoding a D9 fatty acid desaturase homologue was induced specifically in rose petals after anthesis [28], indicating that fatty acid metabolism plays an important role during rose flower development. It has been suggested
* Corresponding author. Tel: +81-75-9628807; fax: + 81-75-9628262.
E-mail address: Masako –[email protected] (M.
Fukuchi-Mizutani).
that fatty acid hydroxyperoxides and their metabolites play some role in flower senescence and the activity of lipoxygenase, the key enzyme of hydroxyperoxidation of fatty acids, is elevated during senescence of carnation flowers [22]. Methyl jasmonate, a product of the octadecanoid pathway from the fatty acid hydroperoxide, has been shown to promote senescence of petunia and dendrobium flowers [29]. Therefore, we decided to investigate whether a lipoxygenase was involved in the senescence of rose flowers.
In this study, we have isolated a cDNA encod-ing a lipoxygenase isoform and found that this cDNA is expressed in rose petals in response to senescence.
2. Materials and methods
2.1. Plant materials
Petals of Rosa hybridacv. Kardinal (grown in a green house in Victoria, Australia) were harvested from flowers at different developmental stages defined as follows: stage 1, closed and pigmenting buds; stage 2, buds still closed and heavily pig-mented; stage 3, flowers whose outer petals were just opening. Flower stems were harvested at stage 3 and the bottom half of the cut stems were kept in distilled water at room temperature; stage 4, flowers 2 days after harvest; stage 5, flowers 4 days after harvest; stage 6, flowers 6 days after harvest; stage 7, flowers 8 days after harvest. Each of the floral organs as well as young red leaves and mature green leaves were obtained from flower stems on the day of harvest.
2.2. Ethylene treatment
For the treatment of rose flowers with ethylene, excised stems with flowers were kept for 24, 48 or 72 h in an 850 ml chamber with a solution of 0.25M Tris – HCl (pH 8.0) and 0.4% Ethrel (May and Baker Rural Pty., Sydney, NSW, Australia; the concentration of ethephon, a precursor to ethylene, is 480 g/l in Ethrel). Under these condi-tions, the concentration of ethylene in the chamber was calculated to be 650 ppm, as described previ-ously [30]. As a control, excised flower stems were incubated under the same conditions in the ab-sence of Ethrel.
2.3. Isolation and molecular analysis of rose lipoxygenase cDNAs
Total RNA was prepared by differential precipi-tation using 2-butoxyethanol [31]. A cDNA library was constructed from polyA+RNAs isolated from
petals at stage 5 (see above) as described previ-ously [28].
DNA fragments from two tomato lipoxygenase cDNAs, tomloxA and tomloxB [19], were ob-tained by PCR using oligonucleotide primers cor-responding to their 5% and 3% sequences, using a tomato fruit cDNA library (breaker stage, Clon-tech, Inc. Palo Alto, California, USA) as a tem-plate. The amplified fragments were labeled with digoxigenin-dUTP (DIG-dUTP) using a DIG la-beling system (Boehringer, Heidelberg, Germany) and used to screen the rose petal cDNA library. The screening procedures were essentially the same as described previously [32] except for the strin-gency of screening conditions; the hybridization solution contained 30% formamide, and washing was performed at 55°C with 5×SSC (0.75 M NaCl, 0.75 M sodium citrate) containing 1% SDS. The sequences of the cDNAs were determined and analyzed as described previously [32]. The possible localization of the rose lipoxygenase was predicted by a PSORT protein sorting analysis program (National Institute for Basic Biology, Okazaki, Japan, E-mail: psort@nibb. ac. jp).
2.4. Expression of a rose lipoxygenase cDNA in E.coli
The expression vector pTrc99A (Pharmacia Bio-tech, Uppsala, Sweden), was used to express a rose lipoxygenase cDNA (Rlox1) in E. coli strain JM109. The 5%-half portion of the Rlox1 cDNA
(146 bp in length) was amplified by PCR so that a NcoI site was created just before the putative initiation codon in the Rlox1 cDNA. The am-plified fragment and the remaining 3%-half portion
7.0) broth containing 50 mg/l ampicillin at 37°C. The expression of Rlox1 cDNA was induced by addition of 2 mM isopropyl b-D-thio-galactoside (final concentration), after which the cells were grown for 4 h and used for purification of the recombinant Rlox1 protein.
2.5. Partial purification of the recombinant lipoxygenase protein
The following purification procedures were per-formed at 4°C. The E.coli cells expressing the recombinant Rlox1 protein (64 g, wet wt) were harvested by centrifugation and were ground with 128 g of levigated alminum oxide in a mortar chilled on ice. The homogenate was then sus-pended in 0.01M potassium phosphate buffer, pH 7.2, containing 0.25 M sucrose (buffer A) followed by centrifugation at 8000g for 15 min. The super-natant was dialyzed against buffer A, and applied to a column of Sepabeads FPDA13 (400 ml, Mit-subishi Chemical Industries, Tokyo) equilibrated with buffer A. The lipoxygenase activity was eluted by a linear gradient of 0 – 1.0 M NaCl in buffer A (1.01 each). The active fractions were dialyzed extensively against buffer A followed by Fast Protein Liqid Chromatography (FPLC) with a HiPrep-Q (16/10) column (Pharmacia) equili-brated with buffer A. After extensive washing of the column with buffer A, lipoxygenase activity was eluted with a linear gradient of 0 – 1.0 M NaCl in Buffer A. The active fractions were dialyzed against buffer A and rechromatographed under the same FPLC conditions as described above except that a linear gradient of 0 – 0.5 M NaCl was used. The active fractions were concentrated using an Amicon 8200 ultrafiltration unit with a PM 10 membrane. The concentrate was subject to gel filtration chromatography on Ultrogel AcA44 (Pharmacia, 2×64 cm) equilibrated with buffer A containing 0.2 M NaCl. The active fractions (pu-rity was 20% as judged by SDS-PAGE) were used for further characterization.
2.6. Assay of lipoxygenase acti6ity
2.6.1. Method I
Lipoxygenase activity was assayed by a conju-gated diene method [33]. The reaction mixture (1.0 ml) consisted of 0.1 M sodium acetate buffer, pH 5.0, 0.0025% Tween 20, and 0.2 mM linoleic acid
(final concentrations). After incubation of the mix-ture at 37°C, the reaction was started by adding enzyme (up to 30 ml) and the increase in ab-sorbance at 234 nm was monitored with a Shi-madzu UV-160 spectrophotometer (kinetic mode) with a temperature-controlled cell positioner CPS-240A. The extinction coefficient for the conjugated diene of 25 000M−1cm−1[2] was used for the unit
calculation.
2.6.2. Method II
Formation of lipid hydroperoxides was moni-tored by a visible-spectrophotomeric method as described previously [34].
2.7. Characterization of enzymatic acti6ity
For pH – activity profiles, lipoxygenase activity was assayed using method I except that the reac-tion mixture contained 100 mM of one of the following buffers: pH 3.0 – 6.0, sodium acetate; pH 6.0 – 8.0, potassium phosphate; pH 8.0 – 9.0, Tris-HCl; and pH 8.0 – 10.0, sodium borate.
For temperature – activity profiles, lipoxygenase activity was assayed at various temperatures indi-cated in Fig. 2B, essentially using assay method I. For pH stability, the enzyme was incubated at 4°C in the following buffers (final concentration, 50 mM): pH 3.0 – 6.0, sodium acetate; pH 6.0 – 8.0, potassium phosphate, pH 8.0 – 9.0, Tris – HCl; and pH 8.0 – 10.0, sodium borate. After incubation for 24 h or 3 weeks, 60ml of each enzyme solution was withdraw and assayed for remaining activity by using method I.
2.8. Northern blot analysis
The procedure for Northern blot analysis is essentially the same as described previously [28]. EcoRI-digested Rlox1 cDNA fragment, 0.25 kb, was used as a probe for Northern blot analysis. This fragment contained all of the 3%-noncoding region of Rlox1 together with a 24 bp 3%-terminal
sequence from the coding region.
3. Results and discussion
3.1. Isolation of rose cDNAs
of a lipoxygenase pathway in flower senescence [22,29] led us to analyze the structure and expres-sion of a lipoxygenase gene, which is potentially involved in the mechanism of senescence of rose petals.
Because senescence of rose flowers usually
be-gins to be visible 4 – 5 days after harvest, we used petals at stage 4 (see Section 2.1) as a source of polyA+ RNAs for the construction of a cDNA
library of senescing rose petals. The 3×105
plaques of a rose senescent petal cDNA library were screened with a mixture of two tomato lipoxygenase cDNA fragments, tomlox A and tomlox B [19], amplified by PCR. Four positive clones were obtained and the clone which had the longest inserts of the four clones, termed Rlox1, was used for further characterization.
3.2. Structural analysis of Rlox1 cDNA
The entire sequence of the Rlox1 cDNA is shown in Fig. 1 with the deduced amino acid sequence. The Rlox1 cDNA was 2899 bp in length and contained an open reading frame consisting of 862 amino acids corresponding to the 78 – 2663rd nucleotides of the predicted Rlox1 sequence. Molecular mass and isoelectoric point of the pre-dicted Rlox1 protein were caluculated to be 97 and 5.1 kDa, respectively. The deduced amino acid sequence of the Rlox1 cDNA was highly ho-mologous to those of other plant lipoxygenases. The highest identity, 74%, was found with potato lipoxygenase 3 [35]. The amino acid sequences of the Rlox1 protein contain the C-terminal amino acid sequence, KGIPNSVSI, which is highly con-served among plant lipoxygenases. The Rlox1 protein also contained a sequence of 38 amino acid residues which is conserved in all lipoxyge-nases and contains amino acid residues essential for catalytic function of lipoxygenases [36]. The amino acid residues essential as ligands to the iron cofactor, His-499, His-504, His-690, Asn-694, and Ile-839, as reported in soybean lipoxygenase-1 [37] are conserved in the Rlox1 protein. They corre-spond to His-523, His-528, His-714, Asn-718, and Ile-862, respectively.
Plant lipoxygenases are, in most cases, cytosolic enzymes though some have been shown to localize to vacuoles [38], chloroplasts [14,39], lipid bodies [40], and microsomal membranes [22,41]. Protein sorting analysis of the predicted amino acid se-quence of Rlox1 protein using a PSORT program suggested that the Rlox1 protein is most likely localized in the cytosol because it contained nei-ther putative targeting nor retention signals to any organellas.
Fig. 2. Enzymatic activity of recombinant Rlox1 protein. Extract from E.coli strain JM109 transformed with pTrcR-lox1 was subject to lipoxygenase enzyme assay using linoleic acid as a substrate. The activity was determined by measuring the increase of A235 (dA). (A) pH optimum of the
recombi-nant Rlox1 protein. Average value of the increase ofA235per
3.3. Partial purification and characterization of Rlox1 protein expressed in E. coli
In order to analyze the enzymatic properties of the Rlox1 protein, an expression vector, pTrcR-lox1, was constructed and the cDNA was ex-pressed in E.coli. The expression product from pTrcRlox1 was a soluble protein with a molecular mass of 97 kDa as expected from the protein encoded in the Rlox1 cDNA. The recombinant Rlox1 protein in the soluble fraction of E.coli lysate showed a strong lipoxygenase activity as judged by both assay method I and II (data not shown), which confirmed that the Rlox1 cDNA encodes a lipoxygenase isoform in rose petals.
The recombinant lipoxygenase encoded by Rlox1 was partially purified and characterized. The rose lipoxygenase exhibited an exceptionally low optimum pH for activity of around pH 4.8 (Fig. 2A). This is in striking contrast with most plant lipoxygenases which have a pH optimum in the neutral and alkaline regions [42]. Thus, Rlox lipoxygenase can be designated as an ‘‘acidic lipoxygenase’’. Interestingly, however, the enzyme was the most stable around pH 7 and lost its activity at pHs lower than pH 5 (data not shown). So far, the only other known acidic lipoxygenase is a soluble lipoxygenase isoform of carnation petals, which shows a broad pH optimum of pH 4.9 – 5.8 [22]. The carnation acidic lipoxygenase was shown to be expressed predominantly in senescent petals. It is, therefore, noteworthy that the expression of the rose acidic lipoxygenase gene Rlox1 was also dramatically increased during petal senescence (see Section 3.4.). The Rlox1 lipoxygenase showed the highest activity at 50°C (Fig. 2B).
3.4. Senescence-related expression of the rose lipoxygenase gene
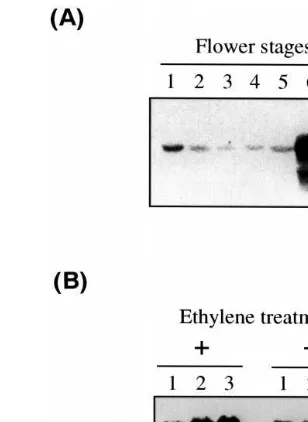
The expression of theRlox1 gene in rose flowers was analyzed by Northern hybridization using a probe derived from the 3%-noncoding sequence of
the Rlox1 cDNA. The transcript of theRlox1 gene was observed in rose petals at all developmental stages and increased dramatically in petals at stages 6 and 7 when petals were withered and discolored (Fig. 3A). We then examined whether the transcription of the Rlox1 gene is enhanced upon application of ethylene which is known to promote flower senescence in plants such as
carna-Fig. 3. Northern blot analysis of senescent-related expression of the Rlox1 gene in rose petals. Ten micrograms of total RNAs were loaded on each lane and probed with Rlox1 cDNA fragment containing 3%noncoding sequence and 24 bp
of coding region. (A) Total RNA from petals at different developmental stages. (B) Total RNA from petals of ethylene-treated flowers (left panel) and non-ethylene-treated flowers (right panel). Flower stems were kept in a chamber for 24 h (lane 1), 48 h (lane 2) and 72 h (lane 3).
tion [24,43,44]. Ethylene treatment of rose flowers caused an increase of theRlox1 transcript to levels more than twice as high as those of non-treated controls (Fig. 3B). The same results were obtained for both senescence- and ethylene-induced expres-sion of the Rlox1 gene when the entire region of the Rlox1 cDNA was used as a probe (data not shown). This result suggests the involvement of Rlox1 lipoxygenase, and so the lipoxygenase path-way, in senescence of rose flowers.
Another possible role of the rose lipoxygenase in petal senescence may be the biosynthesis of jasmonates, which are the end product of the lipoxygenase pathway from a-linolenic acid [3]. Methyl jasmonate has been reported to enhance senescence in petunia and dendrobium flowers via promotion of ethylene production [29]. Therefore, in rose flowers, increased levels of methyl jas-monate resulting from the increased expression of theRlox1 gene may accelerate the process of rose flower senescence.
3.5. Expression of the rose lipoxygenase gene in
6egetati6e and floral organs
The expression of the Rlox1 gene in leaves, stems, and floral organs was analyzed by Northern hybridization using the same probe as described above (Fig. 4a). The transcript of the Rlox1 gene was detected in young leaves and all of the floral organs, but not in stems nor mature leaves. The signal intensity of the detected transcript in young leaves and the floral organs was higher than that in young petals but much lower than in senescent petals. A similar pattern of expression was re-ported for the pea lipoxygenase gene, loxg, whose mRNA is detected mainly in floral tissues and young leaves, but repressed in the subsequent steps of leaf development [47]. These results suggest that the lipoxygenase encoded by Rlox1 is involved in the regulation of leaf development, in addition to a role in promoting petal senescence.
McConn and Browse et.al. [48] indicated that linoleic acid and jasmonate were necessary for pollen development in the triple mutant Arabidop-sis fad3-2 fad7-2 fad8 which contains less than 0.1% trienoic fatty acids. Because jasmonate is synthesized from linoleic acid in the plant lipoxy-genase pathway, this observation indicates a
func-tional significance of a lipoxygenase in pollen development. High expression of the Rlox1 gene in floral organs may also suggest involvement of the Rlox1 lipoxygenase in pollen development.
In conclusion, the Rlox1 cDNA encodes an acidic lipoxygenase which is related to floral and leaf development, especially to petal senescense.
Acknowledgements
The authors would like to thank Dr S. Chandler for his critical reading of the manuscript.
References
[1] B.A. Vick, D.C. Zimmermann, Oxidative systems for modifications of fatty acids: the lipoxygenase pathway, in: P.K. Stumpf, E.E. Conn (Eds.), The Biochemistry of Plants, vol. 9, Academic Press, Orlando, FL, 1987, pp. 53 – 90.
[2] J.N. Siedow, Plant lipoxygenase — structure and func-tion, Annu. Rev. Plant Physiol. Plant Mol. Biol. 42 (1991) 145 – 188.
[3] G. Sembdner, B. Parthier, The biochemistry and the physiological and molecular actions of jasmonates, Annu. Rev. Plant Physiol. Plant Mol. Biol. 44 (1993) 569 – 589.
[4] B.A. Vick, Oxygenated fatty acids of the lipoxygenase pathway, in: T.S. Moore (Ed.), Lipid metabolism in plants, CRC Press, Boca Raton, FL, 1993, pp. 167 – 191. [5] R.A. Creelman, J.E. Mullet, Biosynthesis and action of jasmonates in plants, Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 (1997) 355 – 381.
[6] D.F. Hildebrand, J.G. Rodriguez, C.S. Legg, G.C. Brown, G. Bookjans, The effects of wounding and mite infestation on soybean leaf lipoxygenase levels, Z. Naturforsh. 44c (1989) 655 – 659.
[7] H. Ohta, K. Shida, Y-L. Peng, I. Furusawa, J. Shishiyama, S. Aibara, Y. Morita, A lipoxygenase path-way is activated in rice after infection with the rice blast fungusMagnaporthe grsea, Plant Physiol. 97 (1991) 94 – 98.
[8] E. Koch, B.M. Meier, H-G. Eiben, A. Slusarenko, A lipoxygenase from leaves of tomato (Lycopersicon escu
-lentumMill) is induced in response to plant pathogenic Pseudomonads, Plant Physiol. 99 (1992) 571 – 576. [9] M.L. Fournier, M. Pouenat, H. Rikauer, M.
Ravinovitch-Chable, M.T. Rigaud, M.T. Esquerre-Tu-gaye, Purification and characterization of elicitor-in-duced lipoxygenase in tabacco cells, Plant J. 3 (1993) 63 – 70.
[10] Y. Kondo, Y. Kawai, T. Hayashi, M. Ohnishi, T. Miyazawa, S. Itoh, J. Mizutani, Lipoxygenase in soy-bean seedlings catalyzed the oxygenation of phospho-lipid and such activity changes after treatment with fungal, Biochem. Biophy. Acta. 1170 (1993) 301 – 306. Fig. 4. Northern blot analysis of senescent-related expression
of theRlox1 gene in rose petals. Ten mg of total RNAs were loaded on each lane and probed with Rlox1 cDNA fragment containing 3%noncoding sequence and 24 bp of coding region.
[11] M.A. Melan, X. Dong, M.E. Endara, K.R. Davis, F.M. Ausubel, T.K. Peterman, An Arabidopsis thaliana
lipoxygenase gene can be induced by pathogens, abscisic acid, and methyl jasmonate, Plant Physiol. 101 (1993) 441 – 450.
[12] Y-L. Peng, Y. Shirano, H. Ohta, T. Hibino, K. Tanaka, D. Shibata, A novel lipoxygenase from rice. Primary structure and specific expression upon incompatible in-fection with rice blast fungus, J. Biol. Chem. 269 (1994) 3755 – 3761.
[13] E. Bell, J.E. Mullet, Lipoxygenase gene expression is modulated in plants by water deficit, wounding and methyl jasmonate, Mol. Gen. Genet. 230 (1991) 456 – 462.
[14] E. Bell, J.E. Mullet, Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding, Plant Physiol. 103 (1993) 1133 – 1137. [15] A. Geerts, D. Feltkamp, S. Rosahl, Expression of
lipoxygenase in wounded tubers of Solanum tuberosum
L, Plant Physiol. 105 (1994) 269 – 277.
[16] D.M. Saraviz, J.N. Siedow, The lipoxygenase isozymes in soybean [Glycine max (L) Merr.] leaves. Changes during leaf development, after wounding, and following reproductive sink removal, Plant Physiol. 107 (1995) 535 – 543.
[17] D.M. Saraviz, J.N. Siedow, The differential expression of wound-inducible lipoxygenase genes in soybean leaves, Plant Physiol. 110 (1996) 287 – 299.
[18] F. Mauch, A. Kmecl, U. Schaffrath, S. Volrath, J. Gorlach, E. Ward, J. Ryals, R. Dudler, Mechanosensi-tive expression of a lipoxygenase gene in wheat, Plant Physiol. 114 (1997) 1561 – 1566.
[19] B.J. Ferrie, N. Beaudoin, W. Burkhart, C.G. Bowsher, S. Rothstein, The cloning of two tomato lipoxygenase genes and their differential expression during fruit ripen-ing, Plant Physiol. 106 (1994) 109 – 118.
[20] K.D. Kausch, A.K. Handa, Molecular cloning of a ripening-specific lipoxygenase and its expression during wild-type and mutant tomato fruit development, Plant Physiol. 113 (1997) 1041 – 1050.
[21] T.R. Prakash, P.M. Swamy, P. Suguna, P. Reddanna, Characterization and behavior of 15-lipoxygenase during peanut cotyledonary senescence, Biochem. Biophys. Res. Comm. 172 (1990) 462 – 470.
[22] M-A. Rouet-Mayer, J-M. Bureau, C. Lauriere, Identifi-cation and characterization of lipoxygenase isoforms in senescing carnation petals, Plant Physiol. 98 (1992) 971 – 978.
[23] A. Borochov, W.R. Woodson, Physiology and biochem-istry of flower senescence, Hort. Rev. 11 (1989) 15 – 43. [24] M.S. Reid, M.J. Wu, Ethylene and flower senescence,
Plant Growth Regul. 11 (1992) 37 – 43.
[25] A. Borochov, A.H. Halevy, M. Shinitzky, Increase in microviscosity with aging in protoplast plasmalemma of rose petals, Nature 263 (1976) 158 – 159.
[26] A. Borochov, A.H. Halevy, M. Shinitzky, Senescence and the fluidity of rose petal membranes, Plant Physiol. 69 (1982) 296 – 299.
[27] J.D. Faragher, E. Wachtel, S. Mayak, Changes in the physiol state of membrane lipids during senescence of rose petals, Plant Physiol. 83 (1987) 1037 – 1042.
[28] M. Fukuchi-Mizutani, K. Savin, E. Cornish, Y. Tanaka, T. Ashikari, T. Kusumi, N. Murata, Senescence-induced expression of a homologue of D9 desaturase in rose petals, Plant Mol. Biol. 29 (1995) 627 – 635.
[29] R. Porat, A. Borochov, A.H. Halevy, Enhancement of petunia and dendrobium senescence by jasmonic acid methyl ester is via the promotion of ethylene production, Plant Growth Regul. 13 (1993) 297 – 301.
[30] K.W. Savin, S.C. Baudinette, M.W. Graham, M.Z. Michael, G.D. Nugent, C.Y. Lu, S.F. Chandler, E.C. Cornish, Antisence ACC oxidase RNA delays carnation petal senescence, Colloqium paper of the American Soci-ety of Horticultural Science (1994) 1 – 9.
[31] K. Manning, Isolation of nucleic acids from plants by differential solvent precipitation, Anal. Biochem. 195 (1991) 45 – 50.
[32] M. Fukuchi-Mizutani, Y. Tasaka, Y. Tanaka, T. Ashikari, T. Kusumi, N. Murata, Characterization ofD9 Acyl-lipid desaturase homologues from Arabidopsis thaliana, Plant Cell Physiol. 39 (1998) 247 – 253. [33] S. Grossman, R. Zakut, Determination of the activity of
lipoxygenase (lipoxydase), Methods Biochem. Anal. 25 (1979) 303 – 329.
[34] T. Nakayama, Y. Takeura, T. Ueda, Visible spectropho-tomeric assay, purification, and molecular properties of a lipoxygenase from eggplant (Solanum melongena
Linne) fruits, Biochem. Biophys. Res. Comm. 214 (1995) 1067 – 1072.
[35] M.V. Kolomiets, D.J. Hannapel, R.J. Gladon, Nucle-otide sequence of a cDNA clone for a lipoxygenase from abscisic acid-treated potato leaves, Plant Physiol. 112 (1996) 446.
[36] J. Steczko, G.P. Donoho, J.C. Clemens, J.E. Dixon, B. Axelrod, Conserved histidine residues in soybean lipoxy-genase: functional consequences of their replacement, Biochemistry 31 (1992) 4053 – 4057.
[37] W. Minor, J. Steczko, J.T. Bolin, Z. Otwinowski, B. Axelrod, Crystallographic determination of the active site iron and its ligands in soybean lipoxygenase L-1, Biochemistry 32 (1993) 6320 – 6323.
[38] J.L. Tranbarger, V.R. Franceshi, D.F. Hildebrand, H.D. Grimes, The soybean 94-kilodalton vegitative stroage protein is a lipoxygenase that is localized in paraveinal mesophyll cell vacuoles, Plant Cell 3 (1991) 973 – 987. [39] I. Feussner, H. Kuhn, The lipid body lipoxygenase from
cucumber seedlings exhibits unusual reaction specificity, FEBS lett. 367 (1995) 12 – 14.
[40] I. Feussner, H. Kindl, A lipoxygenase is the main lipid body protein in cucumber and soybean cotyledons dur-ing the stage of triglyceride mobilization, FEBS lett. 298 (1992) 223 – 225.
[41] F. Macri, E. Braidot, E. Petrussa, A. Vianello, Lipoxy-genase activity associated to isolated soybean plasma membranes, Biochim. Biophys. Acta. 1214 (1994) 109 – 114.
[42] H.W. Gardner, Recent investigations into the lipoxyge-nase pathway of plants, Biochim. Biophys. Acta. 1084 (1991) 221 – 239.
[44] M.Z. Michael, K.W. Savin, S.C. Baudinette, M.W. Graham, S.F. Chandler, C-Y. Lu, C. Caesar, I. Gautrais, Y. Young, G.D. Nugent, K.R. Stevenson, E.L.J. O’Connor, C.S. Cobbett, E.C. Cornish, Cloning of ethylene biosynthesis genes involved in petal senes-cence of carnation and petunia, and their antisence expression and molecular aspects of the plant hormone ethylene, in: J.C. Pech, A. Latche, C. Balagne (Eds.), Cellular and Molecular Aspects of the Plant Hormone Ethylene, Kluwer Academic Publishers, Netherlands, 1993, pp. 298 – 303.
[45] M.J. Droillard, M.A. Rouet-Mayer, J.M. Bureau, C. Lauriere, Membrane-associated and soluble
lipoxyge-nase isoform in tomato pericarp. Characterization and involvement on membrane alterations, Plant Physiol. 103 (1993) 1211 – 1219.
[46] D.J. Hardy, M.A.V. Gallegos, J.K. Gaunt, Extracts of
Pisum sati6um metabolise phospholipid via a
lipoxyge-nase-like mechanism, Phytochemistry 30 (1991) 2889 – 2894.
[47] M. Rodriguez-Concepcion, J.P. Beltran, Repression of the pea lipoxygenase geneloxg is associated with carpel development, Plant Mol. Biol. 27 (1995) 887 – 899. [48] M. McConn, J. Browse, The critical requirement for
linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant, Plant Cell 8 (1996) 403 – 416.