Induction of a novel cytochrome P450 (
CYP
96 family) in
periwinkle (
Catharanthus roseus
) cells induced for terpenoid indole
alkaloid production
Audrey Oudin
a, Saı¨d Hamdi
a, Lahzar Oue´lhazi
b, Jean-Claude Che´nieux
a,
Marc Rideau
a,*, Marc Clastre
aaLaboratoire de Biologie Mole´culaire et Biochimie Ve´ge´tale(EA2106),31A6enue Monge,37200Tours,France bUnite´ de Biotechnologie Ve´ge´tale et Resources Ge´ne´tiques,Institute National de Recherche Scientifique et Technique,BP95,
2050Hamman-Lif Tunisia
Received 11 May 1999; received in revised form 19 July 1999; accepted 20 July 1999
Abstract
A full-length cDNA (designatedCDS1) that encodes a cytochrome P450 enzyme (P450) was isolated by combining differential screening and asymmetric PCR amplifications of cDNA libraries from periwinkle (Catharanthus roseus) cell cultures induced for terpenoid indole alkaloid (TIA) synthesis. The deduced polypeptide has 501 aminoacids, a heme-binding cystein domain (the P450 signature), residues conserved in most P450s, and a hydrophobic N-terminal region acting as membrane anchor. CDS1 was assigned as the first member of a new subfamily:CYP96C1. Hybridization of genomic periwinkle DNA with CYP96C1 probe suggested that the protein is encoded by a single-copy gene.CYP96C1 transcripts accumulated only in alkaloid-producing cells showing an association between the induction of the biosynthesis of TIAs and the expression ofCYP96C1. The putative function of the CYP96C1 protein is discussed. © 1999 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:cDNA cloning; Cytochrome P450; Periwinkle (Catharanthus roseus); Terpenoid indole alkaloid (TIA) biosynthesis
www.elsevier.com/locate/plantsci
1. Introduction
The tropical plant Catharanthus roseus (L.) G. Don (Apocynaceae) produces terpenoid indole al-kaloids (TIAs) of high medicinal and economic value such as ajmalicine, catharanthine, vindoline and the bisindoles vinblastine and vincristine. Cell cultures of C. roseus have long been considered to be potential sources of medicinally important
TIAs and various strategies (optimization of medium composition, supply of biosynthetic pre-cursors, treatments with plant growth regulators and elicitors) have been used to increase alkaloid production in suspension-cultured cells [1,2]. How-ever, productivity often remains low and a better understanding of the mechanisms controlling the biosynthesis (and/or accumulation) of TIAs is therefore needed.
The TIA pathway depends on indole and ter-penoid precursors supplied by two convergent branches of the primary metabolite pathways, the shikimate and the isopentenyl diphosphate path-ways. It involves more than twenty enzymes
lo-cated in cytosol, (pro)vacuoles and
chloroplasts/plastids [1]. In the recent years, a few genes encoding enzymes of the TIA pathway have been cloned fromC. roseus, e.g. tryptophan
decar-Abbre6iations: 2,4-D=2,4-dichlorophenoxyacetic acid; FA=fatty acid; G10H=geraniol-10-hydroxylase; IM=induction medium; MM=maintenance medium; pfu=plaque forming unit; PM= pro-duction medium; P450=cytochrome P450 enzyme; TIA=terpenoid indole alkaloid.
The nucleotide sequence data reported in this paper have been deposited in the EMBL database under the following accession number: AJ238402.
* Corresponding author. Tel.: +33-247-367-210; fax: + 33-247-276-660.
E-mail address:[email protected] (M. Rideau)
boxylase, strictosidine synthase, strictosidine glu-cosidase, deacetoxyvindoline 4-hydroxylase [3,4] and more recently the D-1-deoxyxylulose
5-phos-phate synthase (Genbank accession no AJ011840). Other enzymes of the TIA pathway, such as acetyl CoA, deacetylvindoline 4-O-acetyltransferase (fromC.roseus), and isopentenyl diphosphate iso-merase (fromCinchona robusta) were also purified and characterized [5,6]. Some steps of the TIA pathway involve cytochrome P450 (P450)-depen-dent monooxygenases [7,8], a large group of heme-binding containing enzymes which catalyse NADPH- and O2-dependent hydroxylations.
We are currently studying the regulation of TIA production in C. roseus, searching for genes that are preferentially expressed in alkaloid-producing cells. In this paper, we report on the isolation and characterization of a full-length cDNA encoding a new P450, through differential screening of cDNA libraries and asymmetric PCRs. We also showed that the gene is activated by the hormonal signals that lead to TIA accumulation in periwinkle cells.
2. Materials and methods
2.1. Plant material and growth conditions
Periwinkle (C. roseus) cell suspensions (line C20) were maintained on a 7-day growth cycle in the B5 medium of Gamborg [9] supplemented with 58 mM sucrose and 4.5 mM 2,4-dichlorophenoxy-acetic acid (2,4-D) (maintenance medium, MM) [10]. The cells were cultured in 250-ml Erlenmeyer flasks (with 50 ml culture medium) on a rotary shaker (100 rpm) at 24°C in the dark. For experi-mental purposes, 7-day-old cells were subcultured in the following 4 media: the maintenance medium; an ‘inductive medium’ (IM), i.e. MM without 2,4-D; a ‘productive’ medium (PM), i.e. IM to which 5 mM zeatin was added on the third day, and a fourth medium containing both 2,4-D and zeatin (MM+Z). The cells were harvested by vacuum filtration (30 mm nylon cloth) on the fourth and fifth day (for Northern analysis), or on the seventh day (for ajmalicine quantitation). They were weighed (fresh mass), immediately frozen in liquid nitrogen, and either stored at −80°C (Northern and Southern analysis) or freeze-dried (ajmalicine determination).
2.2. Ajmalicine quantitation
50 mg of freeze-dried cells were extracted overnight with 1 ml methanol, then 1 ml of crude extract was spotted onto a silica gel 60 TLC plate (Merck) with the TLC III Autosampler from Ca-mag. The plate was developed for 3 min with ethyl acetate in a horizontal developing chamber, then dried for 1 h at 60°C. Ajmalicine, chosen as a marker of TIA accumulation, was quantified by fluorescence densitometry (TLC Scanner III, l665
nm) [11].
2.3. cDNA libraries
Two periwinkle cDNA libraries were used. The first one is a non-oriented library constructed in 1994. Poly (A+) RNA was isolated from C20 cells
grown in PM, then purified and converted into double-stranded cDNA using the Quick Prep mRNA Purification kit and the Time Saver cDNA synthesis kit (both from Pharmacia). EcoRI –NotI linkers were added to cDNAs ends. After size-fractionation, the cDNA molecules were ligated to
EcoRI-digested dephosphorylated l-ZAPII vector (Stratagene). Recombinant phage DNA was pack-aged with the in vitro Gigapack II Gold Packag-ing Extract (Stratagene). The library was amplified once to yield 2.109 pfu/ml (pfu=plaque forming
units), then stored at −80°C. The second library is an oriented cDNA one, kindly provided in 1998 by Dr Memelink (University of Leiden, the Netherlands).
2.4. Differential screening
80 000 pfu of the non-oriented cDNA library were plated at a density of 5000 pfu per plate (150 mm in diameter). Plaques were transferred to ny-lon filters (Hybond N+, Amersham) and
double-stranded cDNAs were synthesized from poly(A+) RNAs extracted from C20 cells grown in
either MM, IM or PM (+D, −D and +Z
probes, respectively). The probes were [a-32
P]-la-beled with the Multiprime Random Labeling Sys-tem from Amersham. l-ZAP II clones giving stronger hybridization with both the +Z- and the
−D probes than with the +D probe were se-lected, rescued as pBluescript SK plasmids with the in vivo excision system from Stratagene, then sequenced. A partial cDNA that encodes a cy-tochrome P-450 enzyme was further analyzed.
2.5. Isolation of a full-length cDNA encoding a cytochrome P-450 enzyme
By using the l-ZAPII-oriented cDNA library, a
full-length cDNA (designated CDS1 for
C6 atharanthus roseus D6 ifferential S6creening) was generated by asymmetric PCR followed by multi-ple PCRs. Three primers (P1, P2, P3: see Fig. 1) were designed and used (anti-sense orientation) in combination with the universal M13 reverse primer (sense orientation). PCR products am-plified by this primer set would contain the coding information for the N-terminal part of the P450 protein. In a first step, 1ml (1.108pfu) ofl-ZAP II
phage (heat-denatured for 5 min at 94°C) was used as a template for asymmetric PCR with P1 primer only. In a second step, 1ml of the asymmetric PCR mixture was reamplified after addition of the nested P2 primer and the M13 reverse primer. PCR products were resolved by agarose gel elec-trophoresis and the band with approximately the correct size was isolated and purified. In a third step, an aliquot of the purified DNA was amplified with nested P3 primer and M13 reverse primer. Each PCR mixture contained 200 pmol primer(s), 200 mM of each dNTP in 1×buffer containing 3 mM MgCl2 and 2.5 units Taq DNA polymerase
(Promega). The temperature program for all PCRs was 35 cycles of 94°C (30 sec), 56°C (1 min), 72°C (1 min) followed by 10 min at 72°C. Finally, the specific band on agarose gel electrophoresis was isolated and subcloned into pGEM-T vector (Promega). After determining the nucleotide se-quence on both strands, the open reading frame sequence was generated by PCR using an aliquot of cDNA library and PfuDNA polymerase, then, subcloned into pGEM-T vector.
2.6. cDNA sequencing
DNA sequencing was carried out by Genome express SA (France). Search for DNA similarities was performed using BLAST [12] and FASTA [13] algorithms. Prediction of protein-3D structures was based on homologous sequence search using Psi-BLAST algorithm [12,14]. Prediction of protein localization sites in cells was performed using the PSORT computer program [15]. Protein alignment and similarities were determined using the Clustalx multiple sequence alignment software [16].
2.7. Northern blot analysis
Frozen cells (3 g fresh mass) were ground to a fine powder in liquid nitrogen. Total RNA was extracted [17], fractionated on a 1.5% agarose gel containing 2.2 M formaldehyde, capillary trans-ferred to a nylon membrane (Hybond-N+,
Amer-sham), and subsequently baked for 2 h at 80°C. The filter was prehybridized (42°C, 4 h) in buffer (pH 7) containing 6×SSC, 50% formamide, 0.5% SDS, 5×Denhardt’s and 100 mg/ml denatured salmon sperm, then hybridized with the [a-32
P]-dCTP-labeled CDS1 (Megaprime DNA Labeling System, Amersham), using the same solution ex-cept Denhardt’s. The filter was washed twice with 2×SSC, 0.1% SDS at room temperature for 20 min, then twice in 0.1×SSC, 0.1% SDS at 65°C for 10 min, and autoradiographed (XR-Omat LS films). Equal loading of RNA was checked by hybridization of the filter with an [a-32
P]-dCTP-la-beled 18S RNA probe.
2.8. Southern analysis
Genomic DNA was prepared from C20 cells according to [18], digested with PstI and/or BclI, and fractionated on 1% agarose gels. Capillary transfer of DNA fragments, prehybridization, hy-bridization and washings were as above except an higher temperature (65°C) and suppression of for-mamide during hybridization, and an ultimate washing with 0.2×SSC, 0.1% SDS for 10 min at 65°C. As probe, we used a 1113 bp CDS1 insert, obtained after digestion of CDS1 cDNA by PstI. The probe was purified and [a-32P]-dCTP-labeled
3. Results
3.1. Isolation of a gene preferentially expressed in alkaloid-producing cells
The periwinkle line used in the present study accumulates no alkaloid when grown in its mainte-nance medium (MM). Culturing the cells in a 2,4-D-free medium (IM) triggers the production of ajmalicine and other TIAs [10]; adding zeatin at day 3 to the 2,4-D-free medium (PM) further increases the ajmalicine production ([19], and Table 1). To isolate cDNAs that could encode enzymes involved in the regulation of the TIA pathway, we screened a non-oriented cDNA li-brary with +D, −D and +Z probes prepared with mRNAs from cells grown in MM, IM and PM, respectively. A partial cDNA giving higher hybridization with the +Z and −D probes than with the +D probe, was isolated. To generate the 5%-missing region, we took advantage of an ori-ented-cDNA library. A set of three gene-specific primers (Fig. 1) was used together with the M13 reverse universal primer in asymmetric PCRs fol-lowed by a series of PCR reamplifications (see Section 2). The full-length cDNA (designated
CDS1) encoded a putative 501 amino acid-long protein (CDS1, Fig. 1) with a predicted molecular mass of 57.4 kDa.
3.2. The CDS1 cDNA corresponds to the CYP96C1 gene
Search for sequence homologies in databases showed the presence of the signature sequences of cytochrome P450 enzymes [20] in CDS1. In
partic-ular the heme-binding domain (FxxGxxxCxG, Fig. 1) characteristic of such enzymes was found at about 50 aminoacids upstream of the C-termi-nus [21]. Combining data from the hydropathy profile (not shown, [22]) and PSORT analysis sug-gested that the first 24 amino acids formed an endoplasmic reticulum membrane-anchoring se-quence (Fig. 1). The highest homologies between CDS1 and other P450 were found with two P450s from Arabidopsis thaliananamely CYP96A1 (Gen-bank accession no AC002391) and CYP96A2 (Genbank accession no AL021811): 38% and 39% identity, respectively. There are also high homolo-gies with the Arabidopsis thalianaCYP86A1 (Gen-bank accession no X90458, 31% identity, 49% homology), the Vicia sati6a CYP94A1 (Genbank accession no AF030260, 32% identity, 50% homol-ogy) and the Candida maltosa CYP52A2 (Gen-bank accession no M63258, 27% identity, 44% homology) (Fig. 2). According to D.R. Nelson (personal communication), CDS1 must be desig-nated as CYP96C1, the first member of a new subfamily of P450 enzymes. In a second step, we searched for 3-D structure homologies between P450s whose 3-D structures have been published and CYP96C1. We found the highest homologies between CYP96C1 and the bacterial CYP102, in spite of very poor homologies in primary ture. Alignment of CYP96C1 with the 3-D struc-ture of CYP102 showed that the conservation was highest in both the a helices of the heme-binding domain and the a helices preceding this domain ([23], data not shown).
3.3. Molecular organization of the CYP96C1 gene
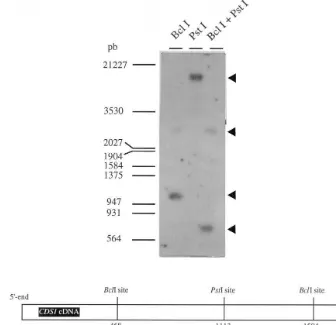
The molecular organization of the CYP96C1 gene was determined by Southern analysis of ge-nomic DNA digested with two restriction endonu-cleases. Digestion with PstI, which does not cut the probe region, resulted in only one hybridiza-tion signal, whereas digeshybridiza-tion with BclI, which cleaved within the probe region, resulted in two signals. This simple hybridization pattern is con-sistent with a single-copy of the CYP96C1 gene (Fig. 3).
3.4. Accumulation of CYP96C1 mRNA in periwinkle cells
C20 cells were grown in MM, IM, PM and also in a fourth medium containing both 2,4-D and
Table 1
Effects of 2,4-D and zeatin (Z) on alkaloid accumulation in
C.roseus cellsa Ajmalicine accu- nd nd 743970
mulationmg/g DM
aThe C20D cells were cultured in 2,4-D-containing medium
Fig. 3. Southern blot analysis ofC.roseusgenomic DNA. DNA (15mg) from C20 cells was totally digested with eitherBclI,PstI and the combination ofBclI andPstI. The blot was hybridized with theCDS1 probe, a 1113 bp-fragment ofCDS1 cDNA. Size markers (bp,HindIII-EcoRI digested lambda-DNA) are indicated on the left.
zeatin (MM+Z). Ajmalicine did not accumulate in the cells grown in MM and MM+Z, but was produced in cells grown in IM and (higher amounts) PM (Table 1). Similarly, CYP96C1 was not expressed in cells grown in MM or MM+Z.
CYP96C1 transcripts accumulated in cells grown in IM and (slightly more) in PM (Fig. 4). The accumulation of CYP96C1 messages was the highest at day 4 in the alkaloid-accumulating cells (Fig. 4) and occurred before the alkaloid accumu-lation that previous studies have shown to begin at day 5 [24]. These data suggested that CYP96C1 expression was associated (directly or indirectly) with the induction of TIA synthesis in C. roseus
cells.
4. Discussion
We report here on the isolation of a cDNA encoding a novel P450 enzyme from periwinkle. Although there is evidence that a large number of P450 genes are expressed in this plant, only few are yet characterized. A nearly full-length cDNA (Cros1), a full-length cDNA (Cros2) and a gene (Cros3) have been isolated [25,26]; they encoded
Fig. 4. Northern blot analysis of RNA fromC.roseus cells. Each lane contains 20mg of total RNA. Cells were grown in four different media (see Table 1), and were harvested at days 4 and 5. The RNA blot was probed with a32P-labeled partial
cDNA of CYP96C1. Hybridization with a [a-32
closely-related proteins that belong to another family (CYP72) of P450s. The partial P450 cDNA clones isolated by Meijer et al. [27] encoded 16 different amino acid sequences that all differ from CDS1. Relationships between P450s are deter-mined on the basis of amino acid sequence iden-tity. Subfamilies of P450 proteins are distinguished when members of one subfamily share less than 55% identity with those of others [28], a criterion verified for CDS1. So, the novel periwinkle P450
protein belongs to a new P450 subfamily
(CYP96C).
Since CYP96C1 transcript levels were higher in alkaloid-producing cells than in non producing-cells (Fig. 4), this expression pattern may reflect some involvement of the encoded protein in the TIA pathway. The biosynthesis of many classes of secondary metabolites requires P450s and key roles for these enzymes have been identified in the biosynthesis of TIAs from periwinkle [7,29,30]. We questioned whetherCYP96C1 encoded an enzyme of the TIA pathway and a first candidate was geraniol-10-hydroxylase (G10H), which catalyzes the conversion of geraniol to its 10-hydroxy derivative [7]. It is now accepted that the biosyn-thesis of the terpenoid precursor, secologanin, is a limiting factor of TIA synthesis [10,31], and G10H is a potential enzyme for regulatory control of the pathway leading to secologanin [32]. To date, the gene encoding the periwinkle G10H has not been characterized, but recently Ohta et al. [33] have isolated a new P450 (CYP76C1) encoding G10H in Arabidopsis thaliana; sequence comparisons showed that CYP96C1 has not enough homolo-gies with CYP76C1 to conclude it encodes G10H. Other steps of the TIA pathway may also require P450s (e.g. [29]), but CYP96C1 involvement in these reactions remains uncertain since the genes encoding these enzymes are not yet characterized. Sequence comparison revealed highest homolo-gies of CYP96C1 with the plant P450s CYP86A1, CYP94A1 (belonging to the CYP89 clan) and the fungal CYP52 family. On a phylogenetic tree [34], they are related and branch close to the mammal P450 CYP4 and the bacterial CYP102. Moreover, the latter showed significant homologies of 3-D structure with CYP96C1 [23]. Since all these P450s have been reported to catalyse the v-hydroxyla-tion of fatty acids (FAs) [23,34 – 36], it is thus possible that CYP96C1 acts similarly. The biologi-cal significance of plant P450 FA hydroxylase
remains to a large extent unknown, although evi-dence of their involvement at different levels of the plant defense reactions has been provided.
In conclusion, we characterized a cDNA encod-ing a novel P450 in C. roseuscells and showed its regulation by the hormonal signals leading to TIA accumulation. What remains to be elucidated is whether CYP96C1 participates to reactions ac-companying TIA production or is directly linked to it.
Acknowledgements
This research was supported by grants from the ‘Ministe`re de l’Education Nationale, de la
Recherche et de la Technologie’ (MENRT,
France). A doctoral fellowship was awarded by the MENRT to A.O. We are grateful to Dr David Nelson for naming the P450 gene family.
References
[1] R. Verpoorte, R. Van der Heijden, J. Schripsema, J.H.C. Hoge, H.J.G. Ten Hoopen, Plant cell biotechnology for the production of alkaloids: present status and prospects, J. Nat. Prod. 56 (1993) 186 – 207.
[2] P.R.H. Moreno, R. Van der Heijden, R. Verpoorte, Effect of terpenoid precursor feeding and elicitation on formation of indole alkaloids in cell suspension cultures ofCatharanthus roseus, Plant Cell Rep. 12 (1993) 702 – 705.
[3] A.H. Meijer, R. Verpoorte, J.H.C. Hoge, Regulation of enzymes and genes involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus, J. Plant Res. 3 (1993) 145 – 164.
[4] F.A. Vazquez-Flota, E. De Carolis, E. Alarco, V. De Luca, Molecular cloning and characterization of desace-toxyvindoline-4-hydroxylase, a 2-oxoglutarate depen-dent-dioxygenase involved in the biosynthesis of vindoline inCatharanthus roseus(L.) G. Don, Plant Mol. Biol. 34 (1997) 935 – 948.
[5] B. St-Pierre, P. Laflamme, A. Alarco, V. De Luca, The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible-for coenzyme A-dependent acyl transfer, Plant J. 14 (1998) 703 – 713.
[6] A.C. Ramos-Valdivia, R. Van der Heijden, R. Ver-poorte, B. Camara, Purification and characterization of two isoforms of isopentenyl-diphosphate isomerase from elicitor-treated Cinchona robusta cells, Eur. J. Biochem. 249 (1997) 161 – 170.
[8] B. St-Pierre, V. De Luca, A cytochrome P450 monooxy-genase catalyses the first step in the conversion of taber-sonine to vindoline inCatharanthus roseus, Plant Physiol. 109 (1995) 131 – 139.
[9] O.L. Gamborg, R.A. Miller, K. Ojima, Nutrient require-ments of suspension cultures of soybean root cells, Exp. Cell Res. 50 (1968) 151 – 158.
[10] J.M. Me´rillon, L. Ouelhazi, P. Doireau, J.C. Che´nieux, M. Rideau, Metabolic changes and alkaloid production in habituated and non-habituated cells of Catharanthus roseusgrown in hormone-free medium. Comparing hor-mone-deprived non-habituated cells with habituated cells, J. Plant Physiol. 134 (1989) 54 – 60.
[11] A. Yahia, C. Kevers, T. Gaspar, J.C. Che´nieux, M. Rideau, J. Cre`che, Cytokinins and ethylene stimulate indole alkaloid accumulation in cell suspension cultures of
Catharanthus roseusby two distinct mechanisms, Plant Sci. 133 (1998) 9 – 15.
[12] S.F. Altschul, T.L. Madden, A.A. Schaffer, J. Zhang, Z. Zhang, W. Miller, D.J. Lipman, Gapped Blast and Psi-Blast: a new generation of protein database search pro-grams, Nucleic Acid Res. 25 (1997) 3389 – 3402. [13] W.R. Pearson, D.J. Lipman, Improved tools for biological
sequence analysis, Proc. Natl. Acad. Sci. USA 85 (1988) 2444 – 2448.
[14] M. Huynen, T. Doerks, F. Eisenhaber, C. Orengo, S. Sunyaev, Y.P. Yuan, P. Bork, Homology-based fold prediction for Mycoplasma genitalium proteins, J. Mol. Biol. 280 (1998) 323 – 326.
[15] K. Nakai, M. Kanehisa, Knowledge base for predicting protein localization sites in eukaryotic cells, Genomics 14 (1992) 897 – 911.
[16] J.D. Thompson, T.J. Gibson, F. Plewniak, F. Jeanmougin, D.G. Higgins, The ClustalX window interface: flexible strategies for multiple sequence alignment aided by quality analysis tools, Nucleic Acids Res. 24 (1997) 4876 – 4882. [17] R. Kay, A. Chan, J. Mac Pherson, Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes, Science 236 (1987) 1299 – 1302.
[18] S.L. Dellaporta, J. Wood, J.B. Hicks, A plant DNA minipreparation, version 2, Plant Mol. Rep. 1 (1983) 19 – 21.
[19] L. Oue´lhazi, M. Filali, A. De´cendit, J.C. Che´nieux, M. Rideau, Differential protein accumulation in zeatin and 2,4-D-treated cells of Catharanthus roseus. Correlation with indole alkaloid biosynthesis, Plant Physiol. Biochem. 31 (1993) 421 – 434.
[20] V.F. Kalb, J.C. Loper, Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity, Proc. Natl. Acad. Sci. USA 85 (1988) 7221 – 7225.
[21] C. Chapple, Molecular-genetic analysis of plant cy-tochrome P450-dependent monooxygenases, Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 (1998) 311 – 343. [22] J. Kyte, R.F. Doolittle, A simple method for displaying
the hydropathic character of a protein, J. Mol. Biol. 157 (1982) 105 – 132.
[23] K.G. Ravichandran, S.S. Boddupalli, C.A. Hasemann, J.A. Peterson, J. Deisenhofer, Crystal structure of hemo-protein domain of P450BM-3, a prototype for microsomal P450s, Science 261 (1993) 731 – 736.
[24] A. De´cendit, D. Liu, L. Oue´lhazi, P. Doireau, J.M. Me´rillon, M. Rideau, Cytokinin-enhanced accumulation of indole alkaloids in Catharanthus roseus cell cultures. The factors affecting the cytokinin response, Plant Cell Rep. 11 (1992) 400 – 403.
[25] H.P. Vetter, U. Mangold, G. Schro¨der, F.J. Marner, D. Werck-Reichhart, J. Schro¨der, Molecular analysis and heterologous expression of an inducible cytochrome P-450 protein from periwinkle (Catharanthus roseus L.), Plant Physiol. 100 (1992) 998 – 1007.
[26] U. Mangold, J. Eichel, A. Batschauer, T. Lanz, T. Kaiser, Gene and cDNA for plant cytochrome P450 proteins (CYP72 family) fromCatharanthus roseusand transgenic expression of the gene and a cDNA in tobacco and
Arabidopsis thaliana, Plant Sci. 96 (1994) 129 – 136. [27] A.H. Meijer, E. Souer, R. Verpoorte, J.H.C. Hoge,
Isolation of cytochrome P-450 cDNA clones from the higher plantCatharanthus roseusby a PCR strategy, Plant Mol. Biol. 22 (1993) 379 – 383.
[28] D.R. Nelson, L. Koymans, T. Kamataki, J.J. Stegeman, R. Feyereisen, D.J. Waxman, M.R. Waterman, O. Gotoh, M.J. Coon, R.W. Estabrook, I.C. Gunsalus, D.W. Nebert, P450 superfamily: update on new sequences, gene map-ping, accession nos and nomenclature, Pharmacogenetics 6 (1996) 1 – 42.
[29] B. St-Pierre, V. De Luca, A cytochrome P-450 monooxy-genase catalyses the first step in the conversion of taber-sonin to vindoline inCatharanthus roseus, Plant Physiol. 109 (1995) 131 – 139.
[30] A. Contin, G. Collu, R. Van der Heijden, R. Verpoorte, The effects of phenobarbital and ketoconazole on the alkaloid biosynthesis inCatharanthus roseus cell suspen-sion cultures, Plant Physiol. Biochem. 37 (1999) 139 – 144. [31] D. Hallard, R. Van der Heijden, R. Verpoorte, M.I. Lopes Cardoso, G. Pasquali, J. Memelink, J.H.C Hoge, Suspen-sion cultured transgenic cells of Nicotiana tabacum ex-pressing tryptophan decarboxylase and strictosidine synthase cDNAs fromCatharanthus roseusproduce stric-tosidine upon secologanin feeding, Plant Cell Rep. 17 (1997) 50 – 54.
[32] O. Schiel, I. Witte, J. Berlin, Geraniol-10-hydroxylase activity and its relation to monoterpene indole alkaloid accumulation in cell suspension cultures ofCatharanthus roseus, Z. Naturforsch 38c (1987) 916 – 922.
[33] D. Ohta, M. Mizutani, Plant geraniol/nerol 10 hydroxy-lase and DNA coding therefor, US Patent 5753507, 1998. [34] I. Benveniste, N. Tijet, F. Adas, G. Philipps, J.P. Salau¨n, F. Durst, CYP86A1 fromArabidopsis thalianaencodes a cytochrome P450-dependent fatty acid omega-hydroxy-lase, Biochem. Biophys. Res. Commun. 243 (1998) 688 – 963.
[35] N. Tijet, C. Helvig, F. Pinot, R. Le Bouquin, A. Lesot, F. Durst, J.P. Salau¨n, I. Benveniste, Functional expression in yeast and characterization of a clofibrate-inducible plant cytochrome P-450 (CYP94A1) involved in cutin monomers synthesis, Biochem. J. 332 (1998) 583 – 589. [36] F. Pinot, I. Benveniste, J-P. Salau¨n, F. Durst, Methyl
jasmonate induces lauric acid (-hydroxylase activity and accumulation of CYP94A1 transcripts but does not affect epoxide hydrolase activities inVicia sati6aseedlings, Plant