PHYLOGENETIC ANALYSIS OF SOME INDO
WEST-PACIFIC GROUPER SUBFAMILY EPINEPHELINAE
(SERRANIDAE) FROM INDONESIAN WATERS
YANTI ARIYANTI
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
STATEMENT LETTER
I hereby declare that thesis entitled
“
Phylogenetic Analysis of Some Indo-
West Pacific Grouper Subfamily Epinephelinae (Serranidae) from Indonesian
Waters
”
is original result of my own research supervised under advisory
committee and has never been submitted in any form at any institution before. All
information from other authors cited here are mentioned in the text and listed in
the reference at the end part of the thesis.
Bogor, October 2015
Yanti Ariyanti
SUMMARY
YANTI ARIYANTI. Pylogenetic Analysis of Some Indo West-Pacific Grouper
Subfamily Epinephelinae (Serranidae) from Indonesian Waters. Supervised by
ACHMAD FARAJALLAH and IRMA SHITA ARLYZA
The serranid Subfamily Epinephelinae comprises about 159 species of marine
fishes in 15 genera,
commonly known as groupers, rockcods, hinds, and
seabasses. These commercially important fishes are bottom-associated which is
found in tropical and subtropical waters. Most species occupies coral reefs, but
some inhabit estuaries or on rocky reefs. Grouper has potential economic value in
fisheries, however, the classification and their evolutionary relationship was often
constrained by the incredible number of species, wide distribution, and lack of
morphological key that
it’s used in classification
. It causes any kind fishes of this
subfamily members were caught in the field are often summed up as groupers.
The Indo-Pacific region has the most diverse types of grouper than the other areas
such as Western Atlantic, Eastern Atlantic, and Eastern Pacific. Therefore, it is
becoming a very interesting to understanding genetic relationship of groupers in
this region particularly in Indonesian water.
In this present work, we have used cytochrome oxidase c subunit 1 (CO1)
gene as a molekular marker in order to investigate the molecular relationship
among some Indo West-Pacific grouper. The specimens were obtained from
various sources including fishing rod, seafood market, and marine fishery station
in several sampling point as follow Aceh (Sumatera), Luwuk (Sulawesi), Kupang
(East Nusa Tenggara), Pangandaran (West Java), Raja Ampat (Papua), Sinjai, and
Selayar Island (South Sulawesi).
Tissue samples were used as the source of DNA
is part of the dorsal muscle, gill and fins tissue. All samples for molecular analysis
were stored in 95% alcohol.
The 26 sequences belonging to 5 generas (
Anyperodon, Cephalopholis,
Epinephelus, Plectropomus, and Variola
) and 14 species of some Indo-West
Pacific grouper from several places in Indonesia were obtained and studied herein.
Several sequences have been submitted in to GenBank. Based on the partial
Cytochorome oxidase subunit 1 and using
Haemulon scuderii
as outgroup, a
molecular phylogenetic tree was constructed by Neighbor-Joining (NJ) method
(Kimura 2-parameter). Appearance of the
Anyperodon
within group of
Epinephelus
(
Epinephelus erythrurus
). Analysis were conducted by combining
morphological and molecular identification (CO1) that showed also within this
report.
This result will be helpful in taxonomy and to understanding phylogenetic
relationship analysis among some West Indo-Pacific grouper in Indonesian
waters.
RINGKASAN
YANTI ARIYANTI. Analisis Filogenetik Beberapa Kerapu Subfamili
Epinephelinae (Serranidae) Indo-Pasifik Barat dari Perairan Indonesia. Dibimbing
oleh ACHMAD FARAJALLAH dan IRMA SHITA ARLYZA
Kelompok ikan serranid dari subfamili Epinephelinae terdiri atas sekitar 159
spesies yang termasuk dalam 15 genera, umumnya dikenal sebagai ikan kerapu,
rockcods, hinds, dan seabasses
. Ikan komersial penting ini merupakan ikan
penghuni dasar perairan yang dapat ditemukan di daerah perairan tropis maupun
subtropis. Sebagian besar spesies ikan ini menghuni habitat terumbu karang,
sedangkan sebagian lainnya hidup di muara atau karang berbatu. Kerapu memiliki
nilai ekonomi tinggi, namun kalsifikasi dan hubungan evolusi mereka seringkali
terkendala oleh jumlah spesies yang luar biasa banyak, distribusi yang luas, serta
kurangnya keahlian dalam identifikasi morfologi. Hal ini menyebabkan apapun
jenis ikan yang tertangkap di lapangan dari anggota subfamili Epinephelinae
seluruhnya disebut sebagai kerapu. Wilayah Indo-Pasifik memiliki jenis ikan
kerapu yang paling beragam daripada daerah lain seperti di Atlantik Barat,
Atlantik Timur, dan Pasifik Timur. Oleh karena itu perairan Indo-Pasifik menjadi
wilayah yang sangat menarik untuk memahami hubungan filogenetik ikan kerapu
khususnya di wilayah perairan Indonesia.
Dalam penelitian ini, digunakan ruas gen sitokrom oksidase c subunit 1 (CO1)
sebagai penanda molekuler untuk menganalisis hubungan antara beberapa jenis
kerapu dari wilayah perairan Indonesia.
Spesimen diperoleh dari berbagai macam
sumber termasuk hasil pancing tradisional, pasar ikan, dan tempat pelelangan ikan
dari beberapa titik sampling yaitu Banda Aceh (Sumatera), Luwuk (Sulawesi),
Kupang (East Nusa Tenggara), Pangandaran (West Java), Raja Ampat (Papua),
Sinjai, and Selayar Island (South Sulawesi).
Jaringan
yang digunakan sebagai
sumber DNA berasal dari otot dorsal, insang, serta sirip
.
Seluruh sampel jaringan
untuk keperluan analisis molekuler disimpan dalam alkohol 95%.
Sebanyak 26 urutan sekuen DNA dari 14 spesies kerapu dari wilayah perairan
Indonesia yang termasuk ke dalam 5 genera (
Anyperodon, Cephalopholis,
Epinephelus, Plectropomus,
dan
Variola
) berhasil diperoleh dan dipelajari
hubungan kekerabatannya dalam penelitian ini. Beberapa sekuen juga telah
didepositkan ke GenBank. Sebuah pohon filogeni yang dibangun menggunakan
metode Neighbor-Joining (NJ) (Kimura 2-parameter) berhasil diperoleh
berdasarkan runutan parsial gen CO1 dengan menggunakan
Haemulon scuderii
sebagai outgroup. Kemunculan
Anyperodon
dalam kelompok
Epinephelus
menyebabkan kelompok ini menjadi tidak monofiletik. Genus
Cephalopholis
lebih primitif dibandingkan genus
Epinephelus
.
Plectropomus
dan
Variola
identifikasi berdasarkan pola warna dan beberapa karakter morfologi tersebut
seringkali terkendala oleh adanya variasi intraspesifik serta perbedaan morfologi
yang sangat mencolok pada satu spesies yang sama antara individu yang masih
juvenil dengan individu yang telah dewasa sehingga menimbulkan kebingungan
dalam penentuan spesies kelompok ikan serranid. Berkaitan dengan hal tersebut,
salah satu contoh kasus yang diungkap dalam penelitian ini adalah penentuan
spesies dari individu yang masih juvenile pada genus
Epinephelus
(
Epinephelus
erythtrurus)
. Analisis dilakukan dengan mengombinasikan identifikasi secara
morfologi dan secara molekuler (CO1).
Hasil penelitian ini diharapkan dapat berguna untuk dunia taksonomi dalam
memahami analisis hubungan filogenetik antara beberapa jenis kerapu Indo
Pasifik Barat di perairan Indonesia.
Copyright © 2015 Bogor Agricultural University
All rights reserved
It is prohibited to cite all or a part of this thesis without referring to and
mentioning the source. Citation only permitted for the purpose of education,
research, scientific paper, report, or critical writing only; and it does not defame
the name and honour of Bogor Agricultural University.
Thesis
As partial fulfilment of the requirements for a Master Degree in
Animal Biosciences Master Program of Graduate School of
Bogor Agricultural University
PHYLOGENETIC ANALYSIS OF SOME INDO-WEST
PACIFIC GROUPER SUBFAMILY EPINEPHELINAE
(SERRANIDAE) FROM INDONESIAN WATERS
GRADUATE SCHOOL
BOGOR AGRICULTURAL UNIVERSITY
BOGOR
2015
PREFACE
This thesis would not have been possible without the help and support of
many people. I would like to thank my committee supervisors: Dr Ir Achmad
Farajallah M.Si, my advisor, allowed me to pursue my research interests and
provided me with endless support and knowledge throughout my graduate thesis,
to Dr Irma Shita Arlyza for support and a chances how to learn become a real
researcher, and to my examiner Dr. Ir. Mohammad Mukhlis Kamal, M.Sc for a
great discussion. Thanks to many people who help me in collecting samples;
Irwan Hikmawan, Mihwan Sataral, Maulana Syafril Yusuf, Meutya Agustina, and
Vice Tantri. I am deeply indebted to Mrs. Maria Ulfah for all her help that has
been given. Also to my partner in laboratory for their kindness and sincerity; Mrs.
Tini, Mr. Adi Surahman, Sister Sianturi, Asri Febriana, Puji Utari Ardika, Novita
Anggraeni, and Esa Ayu Pratama. Thanks to all people in Zoo Corner and BSH
2013 have I regard as my second family. Lastly, I would like to thank all my
family and I am especially grateful for my parents and the loved ones, who have
always been my biggest fans and have encouraged me by all means to achieve my
success. Above all, I thank God Allah
Subhanahu Wa T
a’ala
for His almighty.
Bogor, October 2015
Yanti Ariyanti
CONTENTS
LIST OF TABLES
xi
LIST OF FIGURES
xi
LIST OF FIGURES
vi
INTRODUCTION
1
Background
1
Research objective
2
MATERIALS AND METHOD
2
Study Site and Time
2
Sample Collection
2
Morphological identification
2
DNA Extraction and PCR Reaction
2
Visualization
3
Data Analysis
4
RESULT AND DISCUSSION
4
Result
4
Discussion
6
CONCLUSION
17
LIST OF REFERENCES
17
APPENDICES
20
LIST OF TABLES
1
Sampling location and GenBank Accession number of species
grouper in this study
3
2
Average nucleotide frequencies of CO1 sequences in present study
4
3
Maximum Likelihood Estimate of Substitution Matrix
5
4
Mean percentage group distance (Kimura 2-parameter)
5
5
Genetic distance of CO1 sequences in present study
9
6
Morphometric comparison of the
E. erythrurus
specimens in
the present study with those in the literature
13
7
Genetic distance of CO1 sequences from Indonesian
E. erythrurus
and reference sequences from GenBank
14
8
DNA Sequences details of
Epinephelus erythrurus
in GenBank
file version
16
LIST OF FIGURES
1 Schematic structure of
E. erythrurus
(107 mm standard length)
6
2 Neighbour-joining tree based on the CO1 nucleotide sequences of the
grouper specimens analysed in the present work and of species from
GenBank.
7
3 Representative illustration congruence between tree topological and shape
types of generas
4 Neighbour-joining tree based on the CO1 nucleotide sequences
8
of the
E. erythrurus
specimens analysed in the present work and of
species from GenBank (without Thai
E. erythrurus
)
15
5 Neighbour-joining tree based on the CO1 nucleotide sequences of the
E. erythrurus
specimens analysed in the present work and of species
from GenBank (with Thai
E. erythrurus
)
15
APPENDICES
1
Juveniles of
E. erythrurus
photograph at present study
20
2
Type shapes of 5 genera sub family Epinephelinae at present study based
on Heemstra and Randall 1993
21
3
List of GenBank Accession number for references sequences in the present
study
22
1
INTRODUCTION
Background
The Serranid subfamily Epinephelinae comprises about 159 species of marine
fishes in 15 genera, commonly known as groupers, rockcods, hinds, and seabasses
(Heemstra and Randall 1993). Various fishes of this family are known as Kerapu
or Belong in Bahasa (Burhanudin
et al
. 1980). Serranidae are demersal fish that
occupies coral reefs and rocky substrates. Most members of Serranidae inhabit sea
water, while others inhabit freshwater in tropical and temperate regions in around
the world (Craig and Hastings 2007).
Indonesia has a vast water areas with coral reefs, so that there are potentially
groupers (Syaifudin
et al.
2007). According to Allen (2000), Indonesia has
become the leading country for endemism and also boast the highest overall
species diversity including subfamily Epinephelinae. The Indo-Pacific region has
the most diverse types of grouper than the other areas such as Western Atlantic,
Eastern Atlantic, and Eastern Pacific. Therefore, it is becoming interesting to
understanding genetic relationship of groupers in this region particularly in
Indonesian water.
Recently, the reef fishes of the family Serranidae especially subfamily
Epinephelinae still continues studied, however, the classification and their
evolutionary relationship is often constrained by the incredible number of species,
wide distribution, and lack of morphological characters
that it’s used in
classification. Phenotypic identification of the grouper commonly based on color
pattern and some morphological characters. The colour pattern is usually
distinctive enough to identify large adult groupers at the species level, but
intra-specific variations in colour pattern exist for each species (
Heemstra and Randall
1993;
Govindaraju and Jayasankar 2004). In many cases, fishes, and especially
their diverse developmental stages, are difficult to identify using morphological
characters (Teletchea 2009). Furthermore, difficulties in reconstructing the
evolutionary relationship among grouper species is due to their homogeneous
morphology (Smith 1971).
In this present work, we have used cytochrome oxidase c subunit 1 (CO1)
gene as a molekular marker in order to investigate the molecular relationship
among some West Indo-Pacific grouper. A partial sequence of the mitochondrial
cytochrome oxidase c subunit 1 (CO1) gene is commonly used as a barcode with a
size of about 650 bp, has been used in several animal taxa such as insects, birds,
and fish (Hebert
et al
. 2007). CO1 gene has been used in rapid analyses for
commercial purposes, especially for the confirmation of fish species (Ward
et al
.
2005; Barber and Boyce 2006; Wong and Hanner 2008; Sachithanandam
et al
.
2012).
2
juvenile individual of
Epinephelus
genus (
Epinephelus erythrurus
) by combining
morphological identification and molecular based analysis (CO1).
Research objective
This study aimed to analyse the molecular characteristics of grouper species
(subfamily Epinephelinae) collected from several major islands in Indonesia and
to understanding phylogenetic relationship analyses among some Indo
West-Pacific grouper in Indonesian waters by mean of phylogenetic tree construction
through the use of partial CO1 gene segment.
MATERIALS AND METHOD
Study Site and Time
The research was conducted in April 2014 - April 2015 in the Laboratory of
Molecular, Research Center for Oceanography, Indonesian Institute of Sciences
(LIPI-P2O)
and the Laboratory of Animal Function and Behaviour, Departement of
Biology.
Sample Collection
Several collection samples of fishes is belonging to Dr. Achmad Farajallah
and Dr. Irma Shita Arlyza (LIPI, Oceanography). The specimens were collected
from 2013-2015 were obtained from various sources including fishing rod,
seafood market, and marine fishery station in several sampling point as follow
Aceh (Sumatera), Luwuk (Sulawesi), Kupang (East Nusa Tenggara), Pangandaran
(West Java), Raja Ampat (Papua), Sinjai, and Selayar Island (South Sulawesi)
(Table 1). All samples for molecular analysis were stored in 95% alcohol. Tissue
samples were used as the source of DNA is part of the dorsal muscle, gill and fins
tissue.
Morphological identification
Phenotypic characterization was conducted using the FAO species catalogue
of groupers of the world. The length and the morphometric parameters (body
shape, colour and the rays of the dorsal fins etc.) was measured and counted by
the calliper,
lup, and counter.
DNA Extraction and PCR Reaction
Total DNA was extracted from ethanol preserved muscle using DNA
Extraction Kit for animal tissue (Qiagen and Geneaid) by following the
manufacturer's protocol.
Approximately 648 bp were amplified from the 5′ region
of the COI gene using combinations of the fish-specific primers FishF1
(5’
-TCAACCAACCACAAAGACATTGGCAC-
3’)
and
FishR1
(5’
-TAGACTTCTGGGTGGCCAAAGAATCA-
3’) described in
Ward
et al
. (2005)
and pair of AF28
2 (5’
-TCTACCAACCACAAAGACATCGG-
3’) and AF283
(5’
TACTTCTGGGTGTCCRAAGAATCA-
3’) with some modification from
Ivanova (2007, FishBol). The 25
μL PCR mixes included 18.75 μL of nuclease
free water, 2.25
μL of 10× PCR buffer, 1.25 μL of MgCl
2, 0.25 μL of each primer
3
Visualization
Separation of the products was performed using two methods. Amplicon was
performed using 1 % agarose gel that run at a voltage of 100 volts for 30 minutes.
Then proceed with Ethidium Bromide and visualized under Ultra Violet light.
While separation of the products using 6% polyacrilamide gel was run at a voltage
of 200 V for 40 min. Visualization was facilitated by silver staining (Byun
et al
.
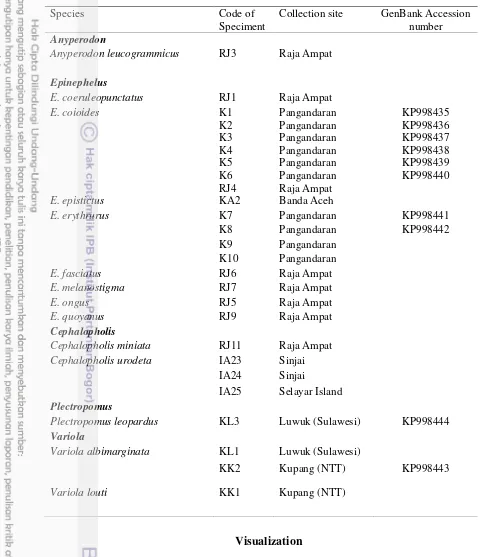
Table 1 Sampling location and GenBank Accession number of species grouper in this study
Species
Code of
Speciment
Collection site
GenBank Accession
number
Anyperodon
Anyperodon leucogrammicus
RJ3
Raja Ampat
Epinephelus
E. coeruleopunctatus
RJ1
Raja Ampat
E. coioides
K1
Pangandaran
KP998435
K2
Pangandaran
KP998436
K3
Pangandaran
KP998437
K4
Pangandaran
KP998438
K5
Pangandaran
KP998439
K6
Pangandaran
KP998440
RJ4
Raja Ampat
E. epistictus
KA2
Banda Aceh
E. erythrurus
K7
Pangandaran
KP998441
K8
Pangandaran
KP998442
K9
Pangandaran
K10
Pangandaran
E. fasciatus
RJ6
Raja Ampat
E. melanostigma
RJ7
Raja Ampat
E. ongus
RJ5
Raja Ampat
E. quoyanus
RJ9
Raja Ampat
Cephalopholis
Cephalopholis miniata
RJ11
Raja Ampat
Cephalopholis urodeta
IA23
Sinjai
IA24
Sinjai
IA25
Selayar Island
Plectropomus
Plectropomus leopardus
KL3
Luwuk (Sulawesi)
KP998444
Variola
Variola albimarginata
KL1
Luwuk (Sulawesi)
KK2
Kupang (NTT)
KP998443
4
2009). (Kimura 1980), including genetic distance calculations and
neighbour-joining (NJ) analysis.
Data Analysis
All amplicons were sequenced commercially following the m
anufacturer’s
protocol. The DNA
sequences were proofread, aligned and edited using MEGA6
(Tamura
et al
. 2013) and BioEdit
(Hall 1999). A Kimura 2-parameter metric was
employed for sequence comparisons (Kimura 1980), including genetic distance
calculations and to generate neighbour-joining trees based on the CO1
region,
with node frequencies calculated based on 1000 bootstrap replicates.
RESULT AND DISCUSSION
Result
We obtained 5 genera and 14 species of some Indo West-Pacific grouper from
several places in Indonesia. Cytochrome oxidase subunit 1 (CO1) were partially
sequenced at least 1 specimens for each species. To form the analysis matrix, the
resulting data were combined with the 14 homologous sequences of the grouper
species downloaded from the
GenBank. A total of 47 taxa were used for the
analysis of which 26 amplified and sequenced in this study, while the rest
(including outgroup) are reference sequence acquired from GenBank database.
Sequence characters
The partial CO1 sequences was 520 base pairs (bp) which translated to 173
amino acids with 200 variable sites, and 182 parsimony informative sites. The
frequencies of mean base composition of all codon was 31.4%, 27.2%, 25.1% and
16.3% for thymine, cytosine, arginine, and guanine, respectively. The content of
A+T (56.5%) was higher than that of C+G (43.5%). The three codon position
differed greatly in their base composition. The nucleotides frequencies of base
composition were almost similar for C, A, and G at the first codon positions,
while T was 18%. At the second position T was 43% and at third G was 6.9%. An
anti-guanine bias was observed for the second (G=12.7%) and third codon
(G=6.9%), it exihibited a strong bias anti-G (Table 2). The estimated
transition/tranversion bias (R) was 3.23 (Kimura 2-parameter), which showed that
transition was obviously more than tranversion (Table 3).
Table 2 Average nucleotide frequencies of CO1 sequences in present study
Base content of CO1 (%)
1
st2
nd3
rdT
C
A
G
T
C
A
G
T
C
A
G
5
Phylogenetic relationship
Except the outgroup, the mean percentage divergence among those 5 genera
was 16.3%. The mean percentage group distance between
Epinephelus
and
Anyperodon
was 13.5% as the lowest, while between
Cephalopholis
and
Plectropomus
was 24% as the highest (Table 4). The minimum pairwise
nucleotide divergence value in CO1 among all taxa was 0.081 between
E.
erythrurus
and
E. coeruleopunctatus,
while the maximum pairwise nucleotide
divergence value was 0.262 between
Epinephelus ongus
and
Variola louti
(Table
5).
Based on the partial Cytochorome oxidase subunit 1 and using
Haemulon
scuderii
as outgroup, a molecular phylogenetic tree was constructed by
Neighbor-Joining (NJ) method (Kimura 2-parameter). The values of bootsrap confidence
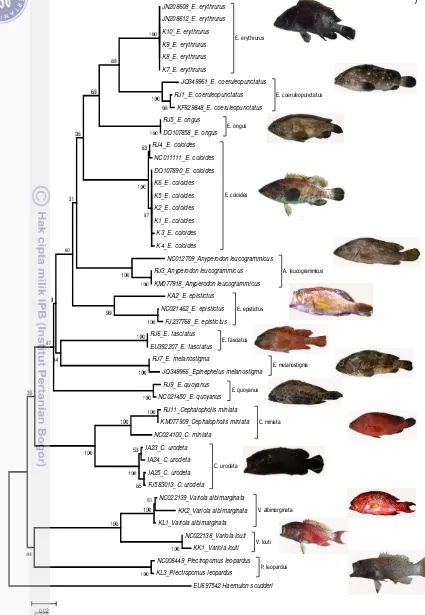
level of nodes were indicated above the branch. Fig. 2 shows that all GenBank
sequences and sequences of subfamily Epinephelinae acquired in this study. NJ
tree clearly exhibited 4 separate groups. The representative illustration congruence
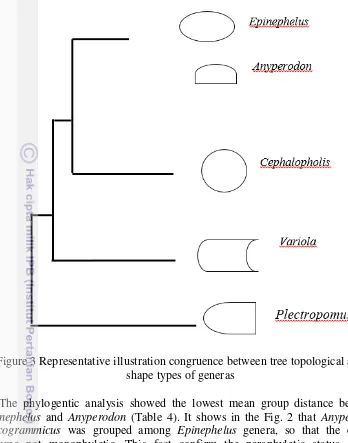
between tree topological and shape types of generas are shown in Fig. 3.
Based on molecular data that the minimum pairwise nucleotide divergence
value in CO1 among all taxa was 0.08 between
E. erythrurus
and
E.
coeruleopunctatus.
According to Noitokr
et al
. (2013), the top ten homologous
analysis results (BLAST) showed that the sequences of
E. erythrurus
were highly
similar to those of
E. coeruleopunctatus
. The other distinctive genus which
comprises two very similar species
V. albimarginata
and
V. louti
has pairwise
nucleotide divergence ranged from 0.09-0.10. Whereas pairwise nucleotide
divergence between
C. urodeta
and
C. miniata
ranged is 0.09.
Epinephelus
cluster
Table 4 Mean percentage group distance (Kimura 2-parameter)
Genera
Mean percentage of group distance (%)
1
2
3
4
5
1.
Epinephelus
-
2.
Anyperodon
13.5
3.
Cephalopholis
18.5
19.1
4.
Variola
21.9
22.1
21.7
5.
Plectropomus
19.9
20.1
24.0
21.6
-
Table 3 Maximum Likelihood Estimate of Substitution Matrix
Original
nucleotide
A
T/U
C
G
A
-
2.96
2.96
19.09
T/U
2.96
-
19.09
2.96
C
2.96
19.09
-
2.96
G
19.09
2.96
2.96
-
6
become not monophyletic because appearance of the
Anyperodon
within the
group.
Plectropomus
and
Variola
stand on the other clade basal position and it is
seem like the primitive group among the subfamily Epinephelinae in this study.
Morphological and molecular based analysis (CO1) identification to confirm
juveniles of Epinephelus erythrurus
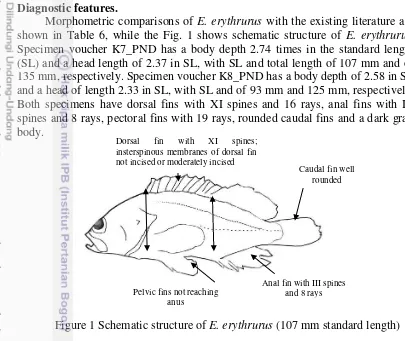
Diagnostic features.
Morphometric comparisons of
E. erythrurus
with the existing literature are
shown in Table 6, while the Fig. 1 shows schematic structure of
E. erythrurus
.
Specimen voucher K7_PND has a body depth 2.74 times in the standard length
(SL) and a head length of 2.37 in SL, with SL and total length of 107 mm and of
135 mm, respectively. Specimen voucher K8_PND has a body depth of 2.58 in SL
and a head of length 2.33 in SL, with SL and of 93 mm and 125 mm, respectively.
Both specimens have dorsal fins with XI spines and 16 rays, anal fins with III
spines and 8 rays, pectoral fins with 19 rays, rounded caudal fins and a dark gray
body.
CO1-based analysis.
The two
E. erythrurus
DNA sequences were submitted individually to
GenBank under accession numbers KP998441 for the K7_PND specimen voucher
and KP998442 for the K8_PND specimen voucher (Table 8). Of the 640
–
651-bp
basic taxonomic sequence length, we were able to obtain 548 bp.
Discussion
Based on sequence character analysis of the CO1 demonstrate that there is
transition and transversion (Table 2). An anti-guanine bias was observed for the
second (G=12.7%) and third codon (G=6.9%) position commonly observed in
fishes (Cantatore
et al
. 1994). As Magio
et al
. (2005) stated that the presence of
compositional bias in every case causes the transition/transversion ratios to vary
for different positions within the codons due to differences in selection pressure,
the third codon position is more likely to be silent than the first and second.
Epinephelinae sequence analysis showed values and characteristic which similar
to data reported on other fishes (Cantatore
et al
. 1994; Ward
et al
. 2005).
Figure 1 Schematic structure of
E. erythrurus
(107 mm standard length)
Caudal fin wellrounded
Anal fin with III spines and 8 rays Pelvic fins not reaching
anus
7
(Photo credits: A. Farajallah, IS. Arlyza, M. Agustina, MS. Yusuf, Y. Ariyanti)
Figure 2 Neighbour-joining tree based on the CO1 nucleotide sequences of the grouper
specimens analysed in the present work and some sequences from GenBank. The
numbers at the nodes indicate bootstrap values for 1000 replicates.
JN208608_E. erythrurus JN208612_E. erythrurus K10_E. erythrurus K9_E. erythrurus K8_E. erythrurus K7_E. erythrurus E. erythrurus JQ349961_E. coeruleopunctatus RJ1_E. coeruleopunctatus KF929848_E. coeruleopunctatus E. coeruleopunctatus RJ5_E. ongus DQ107858_E. ongus E. ongus RJ4_E. coioides NC011111_E. coioides DQ107890_E. coioides K6_E. coioides K5_E. coioides K2_E. coioides K1_E. coioides K3_E. coioides K4_E. coioides E. coioides NC012709_Anyperodon leucogrammicus RJ3_Anyperodon leucogrammicus KM077918_Anyperodon leucogrammicus A. leucogrammicus KA2_E. epistictus NC021462_E. epistictus FJ237768_E. epistictus E. epistictus RJ6_E. fasciatus EU392207_E. fasciatus E. fasciatus RJ7_E. melanostigma JQ349966_Epinephelus melanostigma E. melanostigma RJ9_E. quoyanus NC021450_E. quoyanus E. quoyanus RJ11_Cephalopholis miniata KM077909_Cephalopholis miniata NC024100_C. miniata C. miniata IA23_C. urodeta IA24_C. urodeta IA25_C. urodeta FJ583013_C. urodeta C. urodeta NC022139_Variola albimarginata KK2_Variola albimarginata KL1_Variola albimarginata V. albimarginata NC022138_Variola louti KK1_Variola louti V. louti NC008449_Plectropomus leopardus KL3_Plectropomus leopardus P. leopardus
8
Figure 3 Representative illustration congruence between tree topological and
shape types of generas
The phylogentic analysis showed the lowest mean group distance between
Epinephelus
and
Anyperodon
(Table 4). It shows in the Fig. 2 that
Anyperodon
leucogrammicus
was grouped among
Epinephelus
genera, so that the cluster
become not monophyletic. This fact confirm the paraphyletic status of the
Epinephelus
(Craig
et al.
2001; Zhu
et al
. 2008; You
et al
. 2013). Currrent
classification,
Anyperodon
is distinctive monotypic genus is probably most closely
related to
Epinephelus
, with which it shares XI dorsal-fin spines and the absence
of trisegmental ptetrygiophores, but it differs from
Epinephelus
(and all other
groupers) in its lacking teeth on the palatines.
Anyperodon
is also unique among
groupers in its elongate groupers, but none of these are as compressed as
Anyperodon
(Hemstra & Randall 1993).
The genera position between
Epinephelus
and
Cephalopholis
was also similar
with Craig
et al
(2001), using 16S gene, and then confirmed by Craig and Hasting
(2006) that support the valid genus of the
Cephalopholis
separate from
Epinephelus
.
Cephalopholis
is more primitive than genus
Epinephelus.
Cephalopholis
is one of the largest genera (besides
Mycteroperca
and
9
Table 5
Genetic distance of CO1 sequences in present study (below the diagonal), with standard error estimates (above the diagonal)
Sequence RJ1 RJ5 RJ6 RJ7 RJ9 KA2 RJ4 K1 K2 K3 K4 K5 K6 K7 K8 K9 K10 RJ3 RJ11 IA23 IA24 IA25 KK1 KK2 KL1 KL3
10
Cephalopholis
spp. has often been misidentified as
Epinephelus
spp., for
example, species of
Chepalopholis
have only IX dorsal-fin spines, and
Epinephelus acanthistius
of the Eastern Pacific
also has the same dorsal-fin
spines. According to Hemstra and Randall (1993), another useful generic
character separating both genera is the presence of 3 to 6 trisegmental
pterygiophores in the dorsal fin of
Cephalopholis
species.
In the separate clade, there was genus of
Variola
and
Plectropomus
that
occupied the basal position among all taxa. This molecular analysis concordant
with previous studies that said all genera with VIII-IX spines includes
Aetheloperca, Cephalopholis, Gracila, Paranthias, Plectropomus, Saloptia
, and
Variola
occupy basal positions in both the ML and MP analyses (Craig and
Hasting 2007; Zhu and Yue 2008). Position of
Epinephelus
is located at the top of
the phylogenetic tree indicating that is the most recently diverged species, which
is in concordant with the fact that it is also the most advanced genus in
Epinephelinae (Craig
et al
. 2001; Craig and Hasting 2006, Ding
et al
. 2006).
E.
erythrurus
and
E. coeruleopunctatus
has the lowest value in pairwise nucleotide
divergence (0.079) among all taxa. According to Noitokr
et al
. (2013), the top ten
homologous alignment results in GenBank showed that the sequences of
E.
erythrurus
were highly similar to those of
E. coeruleopunctatus
.
The
E. erythrurus
-
E. coeruleopunctatus
group has
E. ongus
as its sister taxa in
the CO1 tree.
E. quoyanus
is basal to the other species representing a sister taxa.
E. quoyanus
is one of 9 shallow-water coral reef species that have a rounded
caudal fin and close-set dark brown spots with the pale interspaces forming a
network on the body. This reticulated groupers have been much confused in the
literature, and many museum specimen have been misidentified with the other
species such as
E. bilobatus, E. faveatus, E. hexagonatus, E. macrospilos, E.
maculatus, E. melanostigma, E. merra, and E. spilotoceps
(Hemstra and Randall
1993). The representative illustration congruence between tree topological and
shape types of generas are shown in Fig. 3. The shape of grouper illustrated in Fig.
3 which is shows 5 shape types representation to recognize each generas. There
are 5 symbols to illustrate the shape types of grouper:
=
Epinephelus
=
Anyperodon
=
Cephalopholis
=
Variola
= Plectropomus
11
Fujian provinces of China (You
et al
. 2013). Whereas,
E. epistictus
(dotted
grouper) and
E. fasciatus
(black tip grouper) is known from continental localities
in the tropical Indo-West Pacific region.
E. epistictus
probably of some
commercial importance fish, while the
E. fasciatus
is abundance in shallow water.
Morphological and molecular based analysis (CO1) identification to confirm
juveniles of Epinephelus erythrurus
Based on the criteria shown in Table 6, both of our specimens diagnosed as
juvenile
E. erythrurus
. According to Carpenter and Niem (1999), that the female
species become mature at 15 cm of the SL. Also, the adult colour pattern of this
species is usually irregular pale spots and blotches that join randomly to form an
irregular dark reticulum of the background colour. Some specimens, especially
larger ones, are nearly uniform brown or have pale blotches on the body that are
only faintly visible (Randall and Heemstra 1993)
.According to Noitokr
et al
. (2013), the top ten homologous analysis results
(BLAST) showed that the sequences of
E. erythrurus
were highly similar to those
of
E. coeruleopunctatus
. In the current classification an adult of
E.
coeruleopunctatus
has XI dorsal spines and 15 to 17 rays, the third or fourth spine
longest, its length contained 2.7 to 3.6 times in head length; anal fin with III
spines and 8 rays; pectoral fins large and fleshy, with 17-19 rays; caudal fin
rounded. The colour is brownish grey, the body covered with small pale spots
overlain with large pale blotches; oblique black saddle on rear half of peduncle; 4
to 5 indistinct black blotches at baseof dorsal fin, prominent black streak on
maxillary groove. While juveniles (less than 20 cm standard length) dark grey to
black, covered with prominent pupil-size white spots and smaller white dots
(Randall and Heemstra 1993)
.BLAST searches using the two sequences indicated only nine sequences of
E.
erythrurus
from Thailand and Malaysia in GenBank with the query coverage was
91% and the maximum identity was 99
–
100% with existing
E. erythrurus
sequences in GenBank
.
So that, we used those of nine sequences as the references
to construct phylogenetic tree and to understand genetic distances.
Based on the partial CO1 sequences, a molecular phylogenetic tree was
constructed using the neighbour-joining method (Kimura 2-parameter). The
bootstrap confidence values of the nodes are indicated above each branch.
Interestingly, Thai
E. erythrurus
had the smallest number of nucleotides (420 bp).
We constructed two phylogenetic trees, with and without the Thai
E. erythrurus
sequences (Figures 4 and 5). Intra- and inter-specific genetic distances are shown
in Table 7.
Figure 4 show a phylogenetic tree without the Thai
E. erythrurus
sequence.
The two sequences in the present study were grouped with similar Malaysian
E.
erythrurus
sequences. Some
E. coeruleopunctatus
sequences formed a sister
group with
E. erythrurus
, while the other sequences formed a separate clade.
Furthermore, the intra-specific genetic distance of
E. erythrurus
ranged from 0.00
to 0.02 with the same species from other countries, while the inter-specific genetic
distance between
E. erythrurus
and
E. coeruleopunctatus
ranged from 0.01 to
0.07. However, the phylogenetic tree with the first barcode for Thai
E. erythrurus
12
some
E. coeruleopunctatus
from the other separated clades of
E. erythrurus
and
E.
coeruleopunctatus.
It was suggested that
E. erythrurus
and
E. coeruleopunctatus
are not monophyletic.
The genetic distance between pairs of Indonesian
E. erythrurus
and Thai
E.
erythrurus
ranged from 0.066 to 0.068 (Table 7), suggesting two main genotypes
of this species. Based on the two phylogenetic trees, both of our samples were
grouped with the Indonesian and Malaysian
E. erythrurus
that possessed a
genotype different from the Thai
E. erythrurus
.
E. erythrurus
is a fish of minor commercial importance (Heemstra and
Randall 1993) that is often caught with other grouper species. The species is found
in Pakistan, India, Laccadive Island, Sri Lanka, the Gulf of Thailand, Indonesia,
Singapore, Borneo and the Malaysian Peninsula (Heemstra and Randall 1993;
Carpenter and Niem 1999; Allen
et al
. 2003). Based on the FishBase database,
E.
erythrurus
was recorded in Indonesia from Sulawesi to Java. In museums, we
identified RMNH 13525 (Java, Batavia), RMNH 13524 (Surabaya market), SU
61470 (Sangi Island), FMNH 22515-17 (Borneo, Kalimantan, Balikpapan
Harbour), AMS I.19355-039 (Sabah, Sandakan Island), USNM 183241 (North
Borneo) and FMNH 51717. In 2003, Allen and Adrim stated that the distribution
of the species in Indonesia stretched from Sulawesi to Sumatra, and specimens
were stored in the Western Australia Museum. In Thailand, these species was
recorded in a preliminary checklist of coral reef fishes of the Gulf of Thailand
(Satapoomin 2000). Hegde
et al
. (2013) reported that these species included in the
list of the new record along with their habitats from Goa, West coast of India.
Sluka (2013) also stated that
E. erythrurus
was recorded in the three locations
in nearshore rocky or corral habitats of western India, further that report about
these species can be used as an opportunity to remedy it is Data Deficient (DD)
status in IUCN Red List.
The samples in the present study originated from Bojongsalawe Beach
(7
o43’8.31”S 108
o30’11.59”E)
in the Pangandaran district of West Java,
Indonesia. The shoreline of Bojongsalawe Beach directly faces the Indian Ocean.
Due to a lack of data, the species has not yet been assessed for the IUCN Red List
and also is not included in the Catalogue of Life. Another important result from
this research is that the barcode sequence for Indonesian
E. erythrurus
, which was
previously absent from GenBank, is presented here for the first time.
13
Table 6 Morphometric comparison of the E. erythrurus specimens in the present study with those in the literature
Morphometric characters E. erythrurus (KP998441 [K7_PND]) (Present study) E. erythrurus (KP998442 [K8_PND]) (Present study) E. erythrurus
(Hemstra & Randall 1993)
E. erythrurus
(Carpenter & Niem 1998)
E. erythrurus
(Allen et al. 2003)
Body depth 2.74 times in SL 2.58 times in SL 2.8-3.2 times in SL Standard length 107 mm 93 mm 110-280 mm
Total length 135 mm 125 mm - 430 mm To 430 mm
Head length 2.37 times in SL 2.33 times in SL 2.4-2.7 times in SL
Dorsal fin XI XI XI
Dorsal rays 16 16 15 or 17
Anal spines III III III
Anal rays 8 8 8
Caudal fin Rounded Rounded Rounded
Pectoral fins 19 19 17 - 19
Lateral scales series 97 92 92 - 107
Colour Dark gray Dark gray Olive to reddish brown, usually with irregular pale spots and blotches that join randomly to form an irregular dark reticulum of the background colour. Some specimens, especially the larger ones, nearly uniform brown or with the pale blotches on body only faintly visible
Dark gray with irregular pale spots and randomly joined to form maze-like pattern
Geographical distribution Bojongsalawe beach - Pangandaran district, Indonesia
Pakistan, India, Laccadive Island, Sri Lanka, Gulf of Thailand, Indonesia, Singapore, and Borneo
Pakistan, India, Laccadive Is. Sri Lanka, Gulf of Thailand, Indonesia, and Singapore
Pakistan, Laccadive Is. off India to Malaysian Peninsula and W. Indonesia
Habitat Harbours and estuaries with muddy or
silty-sand bottoms
In harbours and estuaries with muddy or silty-sand bottoms
Solitary, turbid harbours and estuaries with mudy or silty-sand bottoms
Depth 1-20
14
Table 7 Genetic distance of CO1 sequences from Indonesian E. erythrurus and reference sequences from GenBank (below the diagonal),
with standard error estimates (above the diagonal)
Accession number K P 9 9 8 4 4 1 K P 9 9 8 4 4 2 JN 2 0 8 6 1 3 JN 2 0 8 6 1 2 JN 2 0 8 6 0 9 JN 2 0 8 6 1 1 JN 2 0 8 6 1 4 JN 2 0 8 6 0 8 JN 2 0 8 6 0 7 JN 2 0 8 6 1 0 JQ 2 6 8 5 7 6 JQ 3 4 9 9 6 1 JQ 3 4 9 9 6 2 JX 6 7 4 9 9 2 JX 6 7 4 9 9 3 JX 6 7 4 9 9 0 JX 6 7 4 9 9 1 K F 9 2 9 8 4 8 JF 4 9 3 4 3 8 JX 0 9 3 9 0 8
KP998441 E. erythrurus (Indonesia) 0.00 0.00 0.00 0.01 0.01 0.01 0.00 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01
KP998442 E. erythrurus (Indonesia) 0.00 0.00 0.00 0.01 0.01 0.01 0.00 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JN208613 E. erythrurus (Malaysia) 0.00 0.01 0.00 0.01 0.01 0.01 0.00 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01
JN208612 E. erythrurus (Malaysia) 0.00 0.00 0.00 0.01 0.01 0.01 0.00 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JN208609 E. erythrurus (Malaysia) 0.01 0.01 0.01 0.01 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01
JN208611 E. erythrurus (Malaysia) 0.01 0.01 0.02 0.01 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JN208614 E. erythrurus (Malaysia) 0.01 0.01 0.02 0.01 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01
JN208608 E. erythrurus (Malaysia) 0.00 0.00 0.01 0.00 0.01 0.01 0.01 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JN208607 E. erythrurus (Malaysia) 0.00 0.01 0.00 0.00 0.01 0.02 0.02 0.01 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01
JN208610 E. erythrurus (Malaysia) 0.00 0.01 0.00 0.00 0.01 0.02 0.02 0.01 0.00 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JQ268576 E. erythrurus (Thailand) 0.07 0.07 0.07 0.07 0.07 0.07 0.07 0.07 0.07 0.07 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.00 0.01
JQ349961 E. coeruleopunctatus 0.07 0.07 0.08 0.07 0.07 0.07 0.07 0.07 0.08 0.08 0.02 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.01 JQ349962 E. coeruleopunctatus 0.06 0.06 0.07 0.06 0.06 0.06 0.06 0.06 0.07 0.07 0.01 0.01 0.01 0.01 0.01 0.01 0.01 0.00 0.01
JX674992 E. coeruleopunctatus 0.02 0.02 0.03 0.02 0.01 0.01 0.01 0.02 0.03 0.03 0.05 0.06 0.05 0.00 0.00 0.00 0.01 0.01 0.01 JX674993 E. coeruleopunctatus 0.02 0.02 0.03 0.02 0.01 0.01 0.01 0.02 0.03 0.03 0.05 0.06 0.05 0.00 0.00 0.00 0.01 0.01 0.01
JX674990 E. coeruleopunctatus 0.02 0.02 0.03 0.02 0.01 0.01 0.01 0.02 0.03 0.03 0.05 0.06 0.05 0.00 0.00 0.00 0.01 0.01 0.01 JX674991 E. coeruleopunctatus 0.02 0.02 0.03 0.02 0.01 0.01 0.01 0.02 0.03 0.03 0.05 0.06 0.05 0.00 0.00 0.00 0.01 0.01 0.01
KF929848 E. coeruleopunctatus 0.06 0.06 0.07 0.06 0.06 0.06 0.06 0.06 0.07 0.07 0.01 0.02 0.01 0.05 0.05 0.05 0.05 0.00 0.01 JF493438 E. coeruleopunctatus 0.06 0.06 0.06 0.06 0.06 0.06 0.06 0.06 0.06 0.06 0.01 0.01 0.00 0.05 0.05 0.05 0.05 0.01 0.01
15
Figure 4 Neighbour-joining tree based on the CO1 nucleotide sequences of
the
E. erythrurus
specimens analysed in the present work and of species from
GenBank (without Thai
E. erythrurus
). The numbers at the nodes indicate
bootstrap values for 1000 replicates.
Figure 5 Neighbour-joining tree based on the CO1 nucleotide sequences of
E. erythrurus
specimens analysed in the present work and of species fr
16
Table 8 DNA Sequences details of Epinephelus erythrurus in GenBank file version
Definition Locus
K7_PND
Epinephelus erythrurus
CO1 gene CDS
K8_PND
Epinephelus erythrurus
CO1 gene CDS
Accession Number
KP998441
KP998442
Submitted Date
VRT 20-Mar-2015
VRT 20-Mar-2015
Classification
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Actinopterygii; Neopterygii; Teleostei; Euteleostei; Neoteleostei;
Acanthomorpha; Eupercaria; Perciformes; Serranoidei; Serranidae;
Epinephelinae; Epinephelini; Epinephelus
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata;
Euteleostomi; Actinopterygii; Neopterygii; Teleostei;
Euteleostei; Neoteleostei; Acanthomorpha; Eupercaria;
Perciformes; Serranoidei; Serranidae;
Epinephelinae; Epinephelini; Epinephelus
17
CONCLUSION
The 26 sequences belonging to 5 genera and 14 grouper species were obtained
and studied herein. DNA analysis based on partial mitochondrial CO1 gene
sequencing successfully identified and confirmed
E. erythrurus
juveniles; these
DNA sequences have been submitted individually to GenBank. Partial sequencing
of the mitochondrial CO1 gene may be used in rapid analyses for commercial
species purposes, especially species identification at various developmental
stages.
For further work we hope to include more specimens per species to reveal any
hidden polyphyletic taxa. Moreover, we propose more species sampling and
molecular genetic analyses to be done on Indonesian grouper, as it will greatly
improve our understanding of the species relationship.
LIST OF REFERENCES
Allen GR, Burhanuddin. 1976.
Anthias hutomoi
, a new species of Serranidae fish
from Indonesia (Perciformes, Serranidae).
Mar Res Indones.
16: 45-50.
Allen GR, Steene R, Humann P, DeLoach N. 2003. Reef fish identification
Tropical Pacific. Singapore (SG): Star Standard Industries Pte Ltd.
Baldwin C.C, Johnson G.D. 1993. Phylogeny of Epinephelinae (Teleostei:
Serranidae).
Bull Mar Sci.
52(1): 240-283.
Barber P, Boyce SL. 2006. Estimating diversity of Indo-Pacific coral reef
stomatopods through DNA barcoding of stomatopod larvae.
Proc R Soc
Lond B
. 273: 2053-2061.
Burhanudin S, Martosewojo, Djamali A.1980. Ikan- ikan demersal di perairan
Teluk Jakarta. Di dalam: Nontji A dan Djamali A, editor. Teluk Jakarta:
Pengkajian Fisika, Kimia, Biologi dan Geologi Tahun 1975-1979. Jakarta
(ID): Lembaga Oseanologi Nasional. LIPI. 337-360.
Byun SO, Fang Q, Zhou H, Hickford JGH. 2009. An effective method for silver
staining DNA in large numbers of polyacrylamide gels.
Anal Biochem.
385:
174-175.
Cantatore P, Roberti M, Pesole G, Ludovico A, Milella F, Gadaletal MN, Saccone
C. 1994. Evolutionary analysis of cytochrome b sequences in some
perciformes: Evidence for a slower rate of evolution than in mammals
. J
Mol Evol
. 39 (6): 589-597.
Carpenter KE, Niem VH. 1999. FAO Species identification guide for fishery
purposes: The Living marine resources of the Western Central Pacific.
Volume 4 Bony fishes part 2 (Mugillidae to Carangidae). Rome (IT): FAO
of the United Nations.
Craig MT, Pondella DJ, Franck JPC, Hafner JC. 2001. On the Status of the
Serranid Fish Genus
Epinephelus
: Evidence for paraphyly Based upon 16S
rDNA sequence.
Mol Phyl Evol.
19: 121-130.
Craig MT, Hastings PA. 2007. Molecular phylogeny of the groupers of the
subfamily Epinephelinae (Serranidae) with a revised classification of the
Epinephelini.
Ichthyol Res
. 54: 1-17.
18
based on 16S rDNA fragment sequences.
Acta Zoologi Sinica
.52 (3):
504-513.
Govindaraju GS, Jayasankar P. 2004. Taxonomic relationship among seven
species of groupers (genus
Epinephelus
; family Serranidae) as revealed by
RAPD fingerprinting.
Mar Biotechnol.
6: 229-237.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010.
New algorithms and methods to estimate Maximum-Likelihood
Phylogenies: Assessing the performance of PhyML 3.0.
Sys Biol
. 59
(3):307-21.
Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and
analysis program for Windows 95/98/NT.
Nucl Acids Symp Ser.
4: 95-98.
Hebert PDN, Ratnasingham S, De Waard JR. 2003. Barcoding animal life:
cytochrome c oxidase subunit 1 divergences among closely related species.
Proc R Soc
Lond
B.
270: 96
–
99.
Heemstra PC, Randall JE. 1993. FAO species catalogue: An annotated and
illustrated catalogue of the grouper, rockcod, hind, coral grouper, and
lyretail species known to date. Volume 16 Groupers of the world (Family
Serranidae, subfamily Epinephelinae). Rome (IT): FAO of the United
Nations.
Hegde MR, Padate VP, Velip DT, Rivonker CU. 2013. An update inventory of
new records of coastal macrofauna along Goa, west coast of India.
Ind J
Geo Mar Sci.
42 (7): 898-902.
Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer
cocktails for fish DNA barcoding.
Mol Ecol Notes.
7: 544-548.
Kimura M. 1980. A simple method for estimating evolutionary rate of base
edsubstitions trough comparative studies of nucleotide sequences.
J Mol
Evol.
16: 111-120.
Maggio T, Andaloro F, Hemida F, Arculeo M. 2005. A molecular analysis of
some Eastern Atlantic grouper from the
Epinephelus
and
Mycteroperca
genus.
J Experiment Mar Biol Ecol
. 321: 83-92.
Noitkr K, Chaveerach A, Pinthong K, Tanomtong A, Sudmoon R, Tanee T. 2013.
RAPD and barcode analyses of groupers of the genus
Epinephelus
.
Gen Mol
Res
. 12 (4): 5721-5732.
Sachithanandam V, Mohan PM, Chaaithanya IK, Dhivya P, Baskara R. 2012.
DNA barcoding, phylogenetic study of
Epinephelus
spp. from Andaman
coastal region, India.
Indian J Geo Mar Sci.
41(3): 203-211.
Satapoomin U. 2000. A preliminary checklist of coral reef fishes of the Gulf of
Thailand, South China Sea.
Raffles B Zool.
48 (1): 31-53.
Sluka RD. 2013. Coastal marine fish biodiversity along the Western coast of
India.
J Threat Taxa
5
(1): 3574-3579.
Smith CL. 1971. A revision of the American groupers:
Epinephelus
and allied
genera.
Bull Am Mus Nat Hist
. 146: 67-241.
Syafiudin M, Aliah RS, Muslim, Sumantadinata K. 2007. Keterkaitan jumlah
induk terhadap frekuensi pemijahan dan produksi telur ikan kerapu tikus
(
Cromileptes altivelis
).
J Akuakul Indones
. 6(2): 191-196.
19
Teletchea F. 2009. Molecular identification methods of fish species: reassessment
and possible applications.
Rev Fish Biol Fisheries.
19: 265-293.
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. 2005. DNA Barcoding of
Australia’s fish species.
Phil Trans
R Soc B.
360: 1847-1857.
Wong EHK, Hanner RH. 2008. DNA barcoding detect market substitution in
North American seafood.
Food Res Int.
41: 828-837.
You X, Shu L, Li S, Chen j, Lou J, Lu J, Mu Q, Bai J, Xia Q, Chen Q
et al.
2013.
Construction of high-density genetic linkage maps for orange-spotted
grouper
Epinephelus coioides
using multiplex shotgun genotyping.
BMC
Genetics.
14: 113.
Zhu S, Yang Y, Zheng W, Shen X, Zou J. 2008. Molecular phylogenetic
relationship of
Epinephelus
base on sequences of mtDNA Cyt b.
Front Biol
China
. 3 (2): 207-212.
Zhu ZY, Yue GH. 2008. The complete mitochondrial genome of red grouper
20
APPENDICES
Appendix 1 Juveniles of
E. erythrurus
photograph
at present study
E. erythrurus
K8_PND (93 mm standard length)
21
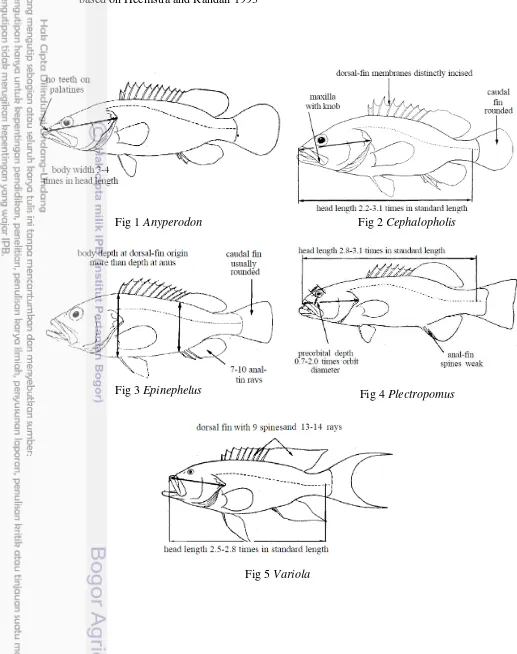
Appendix 2 Type shapes of 5 genera sub family Epinephelinae at present study
based on Heemstra and Randall 1993
Fig 1
Anyperodon
Fig 2
Cephalopholis
Fig 3
Epinephelus
Fig 4
Plectropomus
[image:35.595.34.551.107.761.2]22
Appendix 3 List of GenBank Accession number as references sequences in this
study
No
Species
GenBank Accession number
1
Anyperodon leucogrammicus
NC012709
KM077918
2
E. coeruleopunctatus
JQ349961
KF929848
3
E. coioides
NC011111
DQ107890
NC012709
KM077918
4
E. epistictus
NC021462
FJ237768
5
E. eryhtrurus
JN208608
JN208612
6
E. fasciatus
EU392207
7
E. melanostigma
JQ349966
8
E. ongus
DQ107858
NC012709
9
E. quoyanus
NC021450
10
C. miniata
KM077909
NC024100
11
C. urodeta
FJ583013
12
Plectropomus leopardus
NC008449
13
Variola albimarginata
NC022139
14
Variola louti
NC022138
23
Appendix 4 Alignment of partial CO1 gene sequences in the present study and homologous sequences from GenBank
24
FJ237768_E.epistictus ..A ..C ... ..A ..A ... ..T ... ... ... ..C ..A ... ... ... ... ... ..C ... ... [ 60] DQ107858_E.ongus ..A ... ... ... ... ..T ... ... ... ... ... ..A ... ... ..C ... ..C ... ... ... [ 60] JQ349966_Epinephelus_melanostigma ..A ..C ..G ..C ... ... C.. ... TAT ... ..C ..A ... ... ... ... ... ... ..C ... [ 60] NC021450_E._quoyanus ..A ... T.G ..C ... ..T ..T ... ... ... ... ..A ... ..C ... ..G ... ..G ... ... [ 60] NC011111_E.coioides ..A ... ... ..A ... ... ... ... ... ... ... ..A ... ... ... ... ... ... ... ... [ 60] DQ107890_E.coioides ..A ... ... ..A ... ... ... ... ... ... ... ..A ... ... ... ... ... ... ... ... [ 60] JN208608_E.erythrurus ... ... ... ..A ..T ... ..T ... ... ... ... ..A ... ... ... ... ..C ... ... ... [ 60] JN208612_E.erythrurus ... ... ... ..A ..T ... ..T ... ... ... ... ..A ... ... ... ... ..C ... ... ... [ 60] JQ349961_E._coeruleopunctatus ..T ... ... ... ..T ... ... ... TA. ... ... ... ... ... ... ... ... ... ... ... [ 60] KF929848_E.coeruleopunctatus ... ... ... ... ..T ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [ 60] KM077909_Cephalopholis_miniata ..C ... T.. ..C ... ..T ..T ..A ... ..C ... ..A ... ... ... ... ... ... ..C ... [ 60] NC024100_C._miniata ..C ... T.. ..C ... ..T ..T ..A ... ..C ... ..A ... ... ... ... ... ... ..C ... [ 60] FJ583013_C._urodeta ..T ... T.. ..C ... ..T ..T ..A ... ..C ... ..A ... ... ..G ... ... ... ..C ... [ 60] NC022138_Variola_louti ..A ... ..C T.A ... ..T ... ... ..T ..C ... ..A ..C ... ..T ..C ..C ..G ..C ..G [ 60] NC022139_Variola_albimarginata ... ... ..C T.A ... ..T ... ... ... ... ... ..A ..C ... ..T ..C ..C ..A ..C ..G [ 60] NC008449_Plectropomus_leopardus ..T ... ..T T.A ..A ... ... ..A ... ... ..C ... ... ... ..C ... ..C ..A ... ... [ 60] EU697542_Haemulon_scudderi ..C ..A ..C ..A ..G ... ... ..A ..T ... ..C ..A ... ... ..T ..G ... ..G ..C ... [ 60]
25
K10_E._erythrurus ... ... ... ... ... ... ... ... ... ... ... ... ... ..T ... ... ... ... ... ..G [120] RJ3_Anyperodon_leucogrammicus ... ... ... ... ... ... ..G ... ..T ... ... ... ... ..T ... ... ... ... ..T ... [120] RJ11_Cephalopholis_miniata ... ..T ..C ... ... ... ... ... ... ... ... ... ..G ... ... ..T ..G ... ..T ... [120] IA23_C._urodeta ... ..T ..C ... ... ... ... ... ..T ... ... ... ..G ... ... ... ... ..T ... ... [120] IA24_C._urodeta ... ..T ..C ... ... ... ... ... ..T ... ... ... ..G ... ... ... ... ..T ... ... [120] IA25_C._urodeta ... ..T ..C ... ... ... ... ... ..T ... ... ... ..G ... ... ... ... ..T ..T ... [120] KK1_Variola_louti ..G ..T ..C ... ... ... ..G ... ... ... ..C ... ..G ... ... ... ... ..T ... ..G [120] KK2_Variola_albimarginata ..G ... ..C ... ... ... ..G ... ... ... ... ..C ..G ... ... ... ... ..T ..T .AT [120] KL1_Variola_albimarginata ..G ... ..C ... ... ... ..G ... ... ... ... ..C ..G ... ... ... ... ..T ..T ..T [120] KL3_Plectropomus_leopardus ... ... ... ... ... ... ... ... ... ..A ..C ..A ... ... ... ..T ... ..T ..T ..T [120] NC012709_Anyperodon_leucogrammicus ... ... ... ... ... ... ..G ... ..T ..A ... ... ... ..T ... ... ... ... ..T ... [120] KM077918_Anyperodon_leucogrammicus ... ... ... ... ... ... ..G ... ..T ... ... ... ... ..T ... ... ... ... ..T ... [120] EU392207_E._fasciatus ... ..T ..C ... ... ... ..G ... ... ... ... ..A ... ..T ... ... ... ... ... ... [120] NC021462_E.epistictus ... ..T ..C ... ... ... ... ... ... ... ... ... ..T ..T ... ... ... ..T ... ... [120] FJ237768_E.epistictus ... ..T ..C ... ... ... ... ... ... ... ... ... ..T ..T ... ... ... ..T ... ... [120] DQ107858_E.ongus ... ... ... ... ... ... ... ... ... ... ... ... ... ..T ... ... ... ..T ... ... [120] JQ349966_Epinephelus_melanostigma .A. ..T ..C ... ... ... ..G ... ... ... ... ... ... ... ... ... ... ..T ... ..C [120] NC021450_E._quoyanus ... ..T ... ... ... ... ... ... ... ... ... ... ... ... ... ... ..G ... ..T ..T [120] NC011111_E.coioides ... ... ... ... ... ... ... ... ..T ... ... ... ... ..T ... ... ... ..T ..T ... [120] DQ107890_E.coioides ... ... ... ... ... ... ... ... ..T ... ... ... ... ..T ... ... ... ..T ..T ... [120] JN208608_E.erythrurus ... ... ... ... ... ... ... ... ... ... ... ... ... ..T ... ... ... ... ... ..G [120] JN208612_E.erythrurus ... ... ... ... ... ... ... ... ... ... ... ... ... ..T ... ... ... ... ... ..G [120] JQ349961_E._coeruleopunctatus ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [120] KF929848_E.coeruleopunctatus ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [120] KM077909_Cephalopholis_miniata ... ..T ..C ... ... ... ... ... ... ... ... ... ..G ... ... ..T ..G ... ..T ... [120] NC024100_C._miniata ... ..T ..C ... ... ... ... ... ..T ... ... ... ..G ... ... ... ..G ... ..T ... [120] FJ583013_C._urodeta ... ..T ..C ... ... ... ... ... ..T ... ... ... ..G ... ... ... ... ..T ..T ... [120] NC022138_Variola_louti ..G ..T ..C ... ... ... ..G ... ... ... ..C ... ..G ... ... ... ... ..T ... ..G [120] NC022139_Variola_albimarginata ..G ... ..C ... ... ... ..G ... ... ... ... ..C ..G ... ... ... ... ..T ..T ..T [120] NC008449_Plectropomus_leopardus ... ..T ..C ... ... ... ... ... ... ..A ..C ..A ... ... ... ..T ... ..T ..T ..T [120] EU697542_Haemulon_scudderi ... ..T ..C ... ... ... ..G ... ..T C.C ... ..A ..A ... ... ... ... ... G.. ..C [120]
26
27
JN208608_E.erythrurus ... ... ..T ... ..C ... ..T ..A ... ... ..T ... ... ... ..T ... ... ... ... ... [180] JN208612_E.erythrurus ... ... ..T ... ..C ... ..T ..A ... ... ..T ... ... ... ..T ... ... ... ... ... [180] JQ349961_E._coeruleopunctatus ... ... ..T ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [180] KF929848_E.coeruleopunctatus ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [180] KM077909_Cephalopholis_miniata T.. ... ... ..A ..C ..C ..T ..A ... ... ..T ... ..G ... ..T ... ... ... ..G ..C [180] NC024100_C._miniata ... ... ..T ..G ..C ..C ..T ..A ... ... ..T ... ... ... ..T ... ... ... ..G ..C [180] FJ583013_C._urodeta T.. ... ... ... ... ..C ..T ... ... ... ... ... ..G ... ..T ... ... ..T ... ..C [180] NC022138_Variola_louti T.G ... ..T ..C ... ..T ..T ..A ... ... ..T ... ... ..C ... ..A ... ... ..G ... [180] NC022139_Variola_albimarginata ..G ... ... ..C ... ..T ..T ..A ... ..T ..T ... ... ... ... ..A ... ... ... ... [180] NC008449_Plectropomus_leopardus ... ... ... ..C ..C ... ..T ..A ... ... ..T ... ... ..C ..T ..A ... ... ..G ... [180] EU697542_Haemulon_scudderi ... ..G ..T ..A ..G ..C ... ... ... ... ... ..G ..G ... ... ... ... ..T ... ..C [180]
28
KK1_Variola_louti ... ..T ..T ..T ... T.A ..A ..C ..T ..C ..A ... ..T ... A.G ..A ... ... ... ..A [240] KK2_Variola_albimarginata ... ..T ..T ..T ... T.A ..A ..C ..T ..C ..G ... ..C ... ... ..G ... .G. ... ... [240] KL1_Variola_albimarginata ... ..T ..T ..T ... T.A ..A ..C ..T ..C ..G ... ..C ... ... ..G ... ... ... ... [240] KL3_Plectropomus_leopardus ..A ..T ..T ..T ... ..C ..C ... ..A ..C ..A ..A ..C ..T ... ..A ... ... ..G ..A [240] NC012709_Anyperodon_leucogrammicus ..T ... ... ..T ... ..A ... ..C ..T ... ... ... ..T ... ... ..T ... ... ... ... [240] KM077918_Anyperodon_leucogrammicus ..T ... ... ..T ... ... ... ..C ..T ... ... ... ..T ... ... ... ... ... ... ... [240] EU392207_E._fasciatus ... ..A ... ..T ... ..C ... ... ... ..C ... ..C ..G ... ... ..T ..A ..C ..C ... [240] NC021462_E.epistictus ..T ... ... ..T ... ... ... ... ..T ..C ... ... ..T ..T ... ... ..A ... ... ... [240] FJ237768_E.epistictus ..T ... ... ..T ... S.. ... ... ..T ..C ... ... ..T ..T ... ... ..A ... ... ... [240] DQ107858_E.ongus ... ... ... ..T ... ... ... ... ... ... ... ... ... ... ... ... ... ..C ..C ... [240] JQ349966_Epinephelus_melanostigma ... ... ... ..T ... ... ... ... ..A ..C ... ... ... ... ... ..T ... ... ... ... [240] NC021450_E._quoyanus ... ..T ... ... ... ..A ... ... ..A ... ... ... ... ... ..G ..T ... ..C ... ... [240] NC011111_E.coioides ..T ... ... ... ... ... ... ... ..T ..C ... ... ..T ... ... ... ... ... ..C ... [240] DQ107890_E.coioides ..T ... ... ... ... ... ... ... ..T ..C ... ... ..T ... ... ... ... ... ..C ... [240] JN208608_E.erythrurus ... ... ... ... ... T.A ... ... ... ... ... ... ... ... ... ... ... ... ... ... [240] JN208612_E.erythrurus ... ... ... ... ... T.A ... ... ... ... ... ... ... ... ... ... ... ... ... ... [240] JQ349961_E._coeruleopunctatus ... ... ... ... ... ... ... ... ... ... ... ... ..G ... ... ... ... ... ... ... [240] KF929848_E.coeruleopunctatus ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... ... [240] KM077909_Cephalopholis_miniata ... ..T ... ... ... T.A ... ..G ..A ... ..C ... ... ... ... ... ... ... ... ... [240] NC024100_C._miniata ... ..T ... ... ... T.A ... ..G ..A ... ..C ... ..G ... ... ... ... ... ... ... [240] FJ583013_C._urodeta ... ... ... ... ... ..A ... ..G ..A ..C ..C ... ... ... ... ..T ... ... ... ... [240] NC022138_Variola_louti ... ..T ..T ..T ... T.A ..A ..C ..T ..C ..A ... ..T ... ..G ..A ... ... ... ..A [240] NC022139_Variola_albimarginata ... ..T ..T ..T ... T.A ..A ..C ..T ..C ..G ... ..C ... ... ..G ... ... ... ... [240] NC008449_Plectropomus_leopardus ..A ..T ..T ..T ... ..C ..C ... ..A ..C ..A ..A ..C ..T ... ..A ... ... ..G ..A [240] EU697542_Haemulon_scudderi ... ... ..T ..T ... ..T ..C ... ... ..C ... ..A ... ... ..G ... ..G ... ..G ... [240]
29
30
FJ583013_C._urodeta ... ... ..G ..G ... ... ..T T.. ..T ..T ... ... ... ... ... ... ..C ..C ..T ..T [300] NC022138_Variola_louti ..A ... ... ..C ... ... ... T.G ..A ... ..T T.. ..T ..C ... ... ..T ... ..T ... [300] NC022139_Variola_albimarginata ..A ... ... ..C ... ... ... ..G ..A ..T ..T ... ... ..C ... ... ..T ... ..T ... [300] NC008449_Plectropomus_leopardus ..A ... ... ... ..C ..T ..T ... ..A ... ... ... ... ... ... ... ... ..C ... ..T [300] EU697542_Haemulon_scudderi ..G ... ..T ..C ..C ..T ... T.. ..T ..T ... ... ..G ..C ..C ..A ... ..A ..T ..T [300]
31
EU392207_E._fasciatus ... ..C ... ..C ..A ... ..C T.. ... ... ..T ... ... ... ..G ..G ..T ... ..C ... [360] NC021462_E.epistictus ... ... ..T ... ..C ..G ..C T.. ..C ..A ... ..G ... ... ... ..G ... ..T ..C ... [360] FJ237768_E.epistictus ... ... ..T ... ..C ..G ..C T.. ..C ..A ... ..G ... ... ... ..G ... ..T ..C ... [360] DQ107858_E.ongus ... ..C ... ... ..A T.. ..C T.. ... ... ... ..C ... ... ... ... ... ..T ..C ... [360] JQ349966_Epinephelus_melanostigma ... ..C ... ..C ..A T.. ..C T.G ... ..T ..T ... ... ... ... ..A ... ..T ..C ... [360] NC021450_E._quoyanus ... ...