SUPPLEMENTARY MATERIAL Table 4
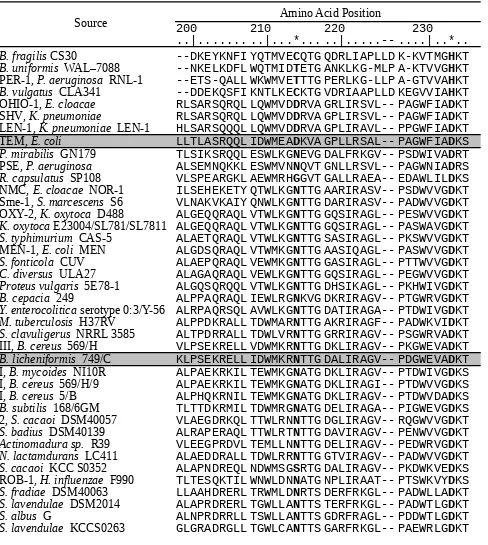
Multiple Amino-Acid Sequence Alignment for 45 Class A -Lactamases (Positions 214 and 233
are marked by asterisks)
Source 200 210 Amino Acid Position220 230
Table 5
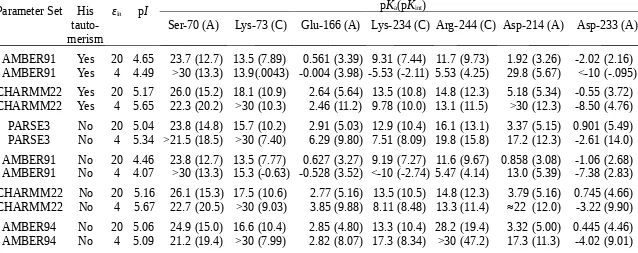
Calculated pKa and pKint for Some Titratable Residues from the Active Site of E. coli TEM-1 (1btl).
Parameter Set His tauto-merism
εin pI pKa(pKint)
Ser-70 (A) Lys-73 (C) Glu-166 (A) Lys-234 (C) Arg-244 (C) Asp-214 (A) Asp-233 (A)
AMBER91 Yes 20 4.65 23.7 (12.7) 13.5 (7.89) 0.561 (3.39) 9.31 (7.44) 11.7 (9.73) 1.92 (3.26) -2.02 (2.16) AMBER91 Yes 4 4.49 >30 (13.3) 13.9(.0043) -0.004 (3.98) -5.53 (-2.11) 5.53 (4.25) 29.8 (5.67) <-10 (-.095) CHARMM22 Yes 20 5.17 26.0 (15.2) 18.1 (10.9) 2.64 (5.64) 13.5 (10.8) 14.8 (12.3) 5.18 (5.34) -0.55 (3.72) CHARMM22 Yes 4 5.65 22.3 (20.2) >30 (10.3) 2.46 (11.2) 9.78 (10.0) 13.1 (11.5) >30 (12.3) -8.50 (4.76) PARSE3 No 20 5.04 23.8 (14.8) 15.7 (10.2) 2.91 (5.03) 12.9 (10.4) 16.1 (13.1) 3.37 (5.15) 0.901 (5.49) PARSE3 No 4 5.34 >21.5 (18.5) >30 (7.40) 6.29 (9.80) 7.51 (8.09) 19.8 (15.8) 17.2 (12.3) -2.61 (14.0) AMBER91 No 20 4.46 23.8 (12.7) 13.5 (7.77) 0.627 (3.27) 9.19 (7.27) 11.6 (9.67) 0.858 (3.08) -1.06 (2.68) AMBER91 No 4 4.07 >30 (13.3) 15.3 (-0.63) -0.528 (3.52) <-10 (-2.74) 5.47 (4.14) 13.0 (5.39) -7.38 (2.83) CHARMM22 No 20 5.16 26.1 (15.3) 17.5 (10.6) 2.77 (5.16) 13.5 (10.5) 14.8 (12.3) 3.79 (5.16) 0.745 (4.66) CHARMM22 No 4 5.67 22.7 (20.5) >30 (9.03) 3.85 (9.88) 8.11 (8.48) 13.3 (11.4) ≈22 (12.0) -3.22 (9.90) AMBER94 No 20 5.06 24.9 (15.0) 16.6 (10.4) 2.85 (4.80) 13.3 (10.4) 28.2 (19.4) 3.32 (5.00) 0.445 (4.46) AMBER94 No 4 5.09 21.2 (19.4) >30 (7.99) 2.82 (8.07) 17.3 (8.34) >30 (47.2) 17.3 (11.3) -4.02 (9.01)
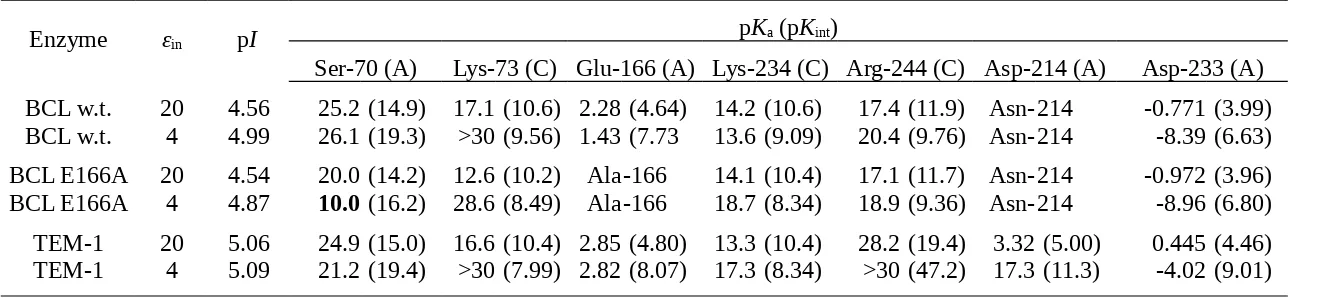
Calculated pKa and pKint for Some Titratable Residues from the Active Site of B. licheniformis 749/C Wild Type (4blm) and Glu166Ala
Mutant (1mbl) Compared with E. coli TEM-1 (1btl).
Enzyme εin pI pKa (pKint)
Ser-70 (A) Lys-73 (C) Glu-166 (A) Lys-234 (C) Arg-244 (C) Asp-214 (A) Asp-233 (A) BCL w.t. 20 4.56 25.2 (14.9) 17.1 (10.6) 2.28 (4.64) 14.2 (10.6) 17.4 (11.9) Asn-214 -0.771 (3.99) BCL w.t. 4 4.99 26.1 (19.3) >30 (9.56) 1.43 (7.73 13.6 (9.09) 20.4 (9.76) Asn-214 -8.39 (6.63) BCL E166A 20 4.54 20.0 (14.2) 12.6 (10.2) Ala-166 14.1 (10.4) 17.1 (11.7) Asn-214 -0.972 (3.96) BCL E166A 4 4.87 10.0 (16.2) 28.6 (8.49) Ala-166 18.7 (8.34) 18.9 (9.36) Asn-214 -8.96 (6.80) TEM-1 20 5.06 24.9 (15.0) 16.6 (10.4) 2.85 (4.80) 13.3 (10.4) 28.2 (19.4) 3.32 (5.00) 0.445 (4.46) TEM-1 4 5.09 21.2 (19.4) >30 (7.99) 2.82 (8.07) 17.3 (8.34) >30 (47.2) 17.3 (11.3) -4.02 (9.01)
[image:4.792.68.735.155.304.2]Table 7
Calculated pKa and pKint for Some Titratable Residues from the Active Site of S. aureus PC1 Wild Type (3blm) and Lys73His Mutant
(1dja) Compared with E. coli TEM-1 (1btl).
Enzyme εin pI
pKa (pKint)
Ser-70 (A) Lys-73(C) Glu-166 (A) Lys-234 (C) Arg-244 (C) Asp-214 (A) Asp-233 (A) SA PC1 20 10.0 24.3 (14.8) 17.9 (10.7) 1.01 (4.50) 14.2 (10.5) >30 (19.1) Asn-214 0.027 (4.66) SA PC1 4 9.52 21.5 (18.9) >30 (9.96) -1.32 (6.92) 13.6 (916) >30 (45.3) Asn-214 -3.22 (9.62)
His-73 (H; H )
SA PC1 K73H 20 ND 25.3 (16.3) 6.87 (6.03; 6.05) 0.614 (6.14) 17.2 (12.4) 17.4 (12.5) Asn-214 1.07 (5.23) SA PC1 K73H 4 ND >30 (23.4) 8.01 (2.08; 3.47) -3.34 (13.2) 26.4 (15.9) 16.9 (10.5) Asn-214 ~-1 (11.4)
Lys-73(C)
TEM-1 20 5.06 24.9 (15.0) 16.6 (10.4) 2.85 (4.80) 13.3 (10.4) 28.2 (19.4) 3.32 (5.00) 0.445 (4.46) TEM-1 4 5.09 21.2 (19.4) >30 (7.99) 2.82 (8.07) 17.3 (8.34) >30 (47.2) 17.3 (11.3) -4.02 (9.01)
Calculated pKa and pKint for Some Titratable Residues from the Active Site of the Model of the Michaelis Complex of E. coli TEM-1,
B. licheniformis 749/C Wild Type, its Glu166Ala Mutant, S. aureus PC1 Wild Type and its Lys73His Mutant with Penicillin G.
Enzyme εin
pKa (pKint)
Ser-70 (A) Lys-73(C) Glu-166 (A) Lys-234 (C) Arg-244 (C) Asp-214 (A) Asp-233 (A)
TEM-1 (91) 20 27.9 (12.4) 16.1 (7.02) -0.662 (2.72) 12.5 (6.91) 13.9 (9.15) 2.05 (3.07) -1.57 (2.57) TEM-1 (91) 4 >28 (13.2) >30 (-5.56) -8.21 (1.96) 3.80 (-6.19) 10.7 (-.35) >17 (5.12) -9.95 (2.03)
TEM-1 20 29.5 (15.4) 17.9 (9.96) 0.785 (4.15) 18.5 (10.9) 23.5 (16.1) 4.93 (5.40) 0.213 (4.59) TEM-1 4 >30 (25.0) >30 (6.35) -1.72 (6.88) >30 (10.2) >30 (29.4) 16.3 (13.2) -4.34 (9.54)
BCL w.t. 20 >30 (15.5) 19.4 (10.6) 0.669 (4.07) 19.5 (10.7) 19.6 (11.7) Asn-214 3.82 (3.48) BCL w.t. 4 >30 (25.0) >30 (9.19) -2.95 (6.23) >30 (9.34) 26.6 (8.60) Asn-214 <-10 (4.36)
BCL E166A mutant 20 26.9 (15.1) 14.7 (10.5) Ala-166 19.6 (10.6) 19.2 (11.8) Asn-214 -2.52 (3.38) BCL E166A mutant 4 >23 (23.4) 22.4 (8.67) Ala-166 >30 (8.74) 26.6 (8.78) Asn-214 <-10 (3.76)
SA w.t. 20 >30 (15.4) 19.8 (10.1) -0.703 (4.14) 19.6 (10.3) 19.7 (11.8) Asn-214 -2.36 (4.03) SA w.t. 4 >30 (24.4) >30 (6.70) <-10 (6.46) >30 (7.46) 23.1 (8.00) Asn-214 <-10 (6.59)
His-73 (H; H)
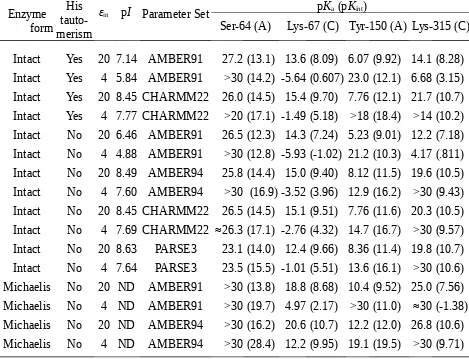
[image:6.792.74.680.170.495.2]Table 9
Calculated pKa and pKint for Some Titratable Residues from the Active Site of the Intact Enzyme P99 E.
cloacae (2blt) and a Model of its Michaelis Complex with Penicillin G.
Enzyme form
His tauto-merism
εin pI Parameter Set pKa (pKint)
Ser-64 (A) Lys-67 (C) Tyr-150 (A) Lys-315 (C)
Intact Yes 20 7.14 AMBER91 27.2 (13.1) 13.6 (8.09) 6.07 (9.92) 14.1 (8.28) Intact Yes 4 5.84 AMBER91 >30 (14.2) -5.64 (0.607) 23.0 (12.1) 6.68 (3.15) Intact Yes 20 8.45 CHARMM22 26.0 (14.5) 15.4 (9.70) 7.76 (12.1) 21.7 (10.7) Intact Yes 4 7.77 CHARMM22 >20 (17.1) -1.49 (5.18) >18 (18.4) >14 (10.2) Intact No 20 6.46 AMBER91 26.5 (12.3) 14.3 (7.24) 5.23 (9.01) 12.2 (7.18) Intact No 4 4.88 AMBER91 >30 (12.8) -5.93 (-1.02) 21.2 (10.3) 4.17 (.811) Intact No 20 8.49 AMBER94 25.8 (14.4) 15.0 (9.40) 8.12 (11.5) 19.6 (10.5) Intact No 4 7.60 AMBER94 >30 (16.9) -3.52 (3.96) 12.9 (16.2) >30 (9.43) Intact No 20 8.45 CHARMM22 26.5 (14.5) 15.1 (9.51) 7.76 (11.6) 20.3 (10.5) Intact No 4 7.69 CHARMM22 ≈26.3 (17.1) -2.76 (4.32) 14.7 (16.7) >30 (9.57) Intact No 20 8.63 PARSE3 23.1 (14.0) 12.4 (9.66) 8.36 (11.4) 19.8 (10.7) Intact No 4 7.64 PARSE3 23.5 (15.5) -1.01 (5.51) 13.6 (16.1) >30 (10.6) Michaelis No 20 ND AMBER91 >30 (13.8) 18.8 (8.68) 10.4 (9.52) 25.0 (7.56) Michaelis No 4 ND AMBER91 >30 (19.7) 4.97 (2.17) >30 (11.0) ≈30 (-1.38) Michaelis No 20 ND AMBER94 >30 (16.2) 20.6 (10.7) 12.2 (12.0) 26.8 (10.6) Michaelis No 4 ND AMBER94 >30 (28.4) 12.2 (9.95) 19.1 (19.5) >30 (9.71)
E. cloacae P99 pI = 8.2; 7.8; 8.3; 8.25; 8.95 (Goering, R. V.; Sanders, C. C.; Sanders, W. E., Jr.; Guay, R.; Guerin, S. Rev. Infect. Dis. 1988, 10, 786-792; Goward, C. R.; Stevens, G. B.; Hammond, P. M.; Scawen, M. D. Chromatogr. 1988, 457, 317-324; Minami, S.; Inoue, M.; Mitsuhashi, S. Antimicrob. Agents. Chemother. 1980, 18, 853-857; Seeberg, A. H.; Tolxdorff-Neutzling, R. M.; Wiedemann, B. Antimicrob. Agents. Chemother. 1983, 23, 918-925.)
Calculated pKa and pKint for Some Titratable Residues from the Active Site of the
DD-Transpeptidase R61 Streptomyces sp. (1cef) and a Model of its Michaelis Complex with
Penicillin G.
Enzyme His tau- to- me-rism
εin pI Parameter
Set
pKa (pKint)
Ser-62 (A) Lys-65 (C) Tyr-159 (A) His-298 (C)
Intact No 20 4.44 AMBER91 ≈29.5 (13.2) 18.1 (8.38) 7.69 (10.2) -1.37 (3.79) Intact No 4 4.02 AMBER91 >30 (14.0) -1.97 (1.71) 27.0 (13.0) <-10 (-3.44) Intact No 20 4.89 CHARMM22 >30 (14.5) 23.6 (10.3) 9.21 (11.6) 2.11 (6.39) Intact No 4 5.22 CHARMM22 >30 (17.0) 13.1 (7.98) 7.84 (17.3) -5.20 (6.92) Intact No 20 4.71 PARSE3 28.3 (14.1) 21.9 (10.3) 9.31 (11.4) 1.56 (6.24) Intact No 4 4.67 PARSE3 18.5 (14.7) 8.55 (8.45) >24 (16.5) -7.08 (5.43) Intact No 20 4.85 AMBER94 ≈30 (14.3) 21.8 (10.1) 9.23 (11.2) 1.38 (5.78) Intact No 4 4.92 AMBER94 27.9 (16.2) 12.9 (7.27) 7.48 (15.1) -6.63 (4.25) Intact Yes 20 ND AMBER94 >30 (16.4) 20.5 (12.6) 9.20 (13.7) 2.27 (7.94;7.26) Intact Yes 4 ND AMBER94 29.8 (21.3) ~10.5 (15.7) 6.92 (22.6) -5.89 (9.72; 8.22)
Michaelis No 20 ND AMBER91 >30 (13.2) 24.7 (8.30) 14.1 (9.82) -3.58 (1.36) Michaelis No 4 ND AMBER91 >30 (16.1) 5.81 (-0.21) >30 (12.6) <-10 (-16.0) Michaelis No 20 ND AMBER94 >30 (14.8) 25.8 (9.99) 13.8 (11.4) -2.32 (3.17) Michaelis No 4 ND AMBER94 >30 (21.0) 20.7 (6.02) >30 (17.3) <-10 (-9.27) Michaelis Yes 20 ND AMBER94 >30 (17.8) 25.9 (12.9) 15.6 (14.2) -0.334 (5.51;6.07) Michaelis Yes 4 ND AMBER94 >30 (33.1) >18 (18.1) >30 (28.4) <-10 (-2.47;1.96)
[image:8.612.77.558.185.577.2]