SUPPLEMENTARY MATERIAL
(Tables S1–S3)
Theoretical Study of Reaction of Trifluoromethyl Radical
with Hydroxyl and Hydrogen Radicals
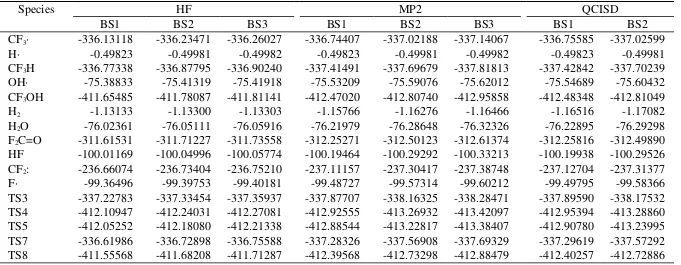
TABLE S1.
The electronic energies calculated by ab initio methods.a
Species HF MP2 QCISD
BS1 BS2 BS3 BS1 BS2 BS3 BS1 BS2
CF3∙ 336.13118 336.23471 336.26027 336.74407 337.02188 337.14067 336.75585 337.02599
H∙ 0.49823 0.49981 0.49982 0.49823 0.49981 0.49982 0.49823 0.49981
CF3H 336.77338 336.87795 336.90240 337.41491 337.69679 337.81813 337.42842 337.70239
OH∙ 75.38833 75.41319 75.41918 75.53209 75.59076 75.62012 75.54689 75.60432
CF3OH 411.65485 411.78087 411.81141 412.47020 412.80740 412.95858 412.48348 412.81049
H2 1.13133 1.13300 1.13303 1.15766 1.16276 1.16466 1.16516 1.17082
H2O 76.02361 76.05111 76.05916 76.21979 76.28648 76.32326 76.22895 76.29298
F2C=O 311.61531 311.71227 311.73558 312.25271 312.50123 312.61374 312.25816 312.49890
HF 100.01169 100.04996 100.05774 100.19464 100.29292 100.33213 100.19938 100.29526
CF2: 236.66074 236.73404 236.75210 237.11157 237.30417 237.38748 237.12704 237.31377
F∙ 99.36496 99.39753 99.40181 99.48727 99.57314 99.60212 99.49795 99.58366
TS3 337.22783 337.33454 337.35937 337.87707 338.16325 338.28471 337.89590 338.17532
TS4 412.10947 412.24031 412.27081 412.92555 413.26932 413.42097 412.95394 413.28860
TS5 412.05252 412.18080 412.21338 412.88544 413.22817 413.38407 412.90780 413.23995
TS7 336.61986 336.72898 336.75588 337.28326 337.56908 337.69329 337.29619 337.57292
TS8 411.55568 411.68208 411.71287 412.39568 412.73298 412.88479 412.40257 412.72886
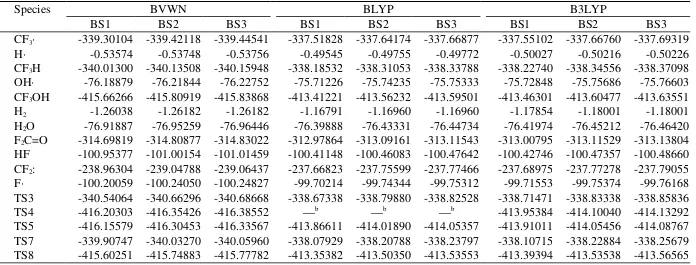
The electronic energies calculated by the DFT methods.a
Species BVWN BLYP B3LYP
BS1 BS2 BS3 BS1 BS2 BS3 BS1 BS2 BS3
CF3∙ 339.30104 339.42118 339.44541 337.51828 337.64174 337.66877 337.55102 337.66760 337.69319
H∙ 0.53574 0.53748 0.53756 0.49545 0.49755 0.49772 0.50027 0.50216 0.50226
CF3H 340.01300 340.13508 340.15948 338.18532 338.31053 338.33788 338.22740 338.34556 338.37098
OH∙ 76.18879 76.21844 76.22752 75.71226 75.74235 75.75333 75.72848 75.75686 75.76603
CF3OH 415.66266 415.80919 415.83868 413.41221 413.56232 413.59501 413.46301 413.60477 413.63551
H2 1.26038 1.26182 1.26182 1.16791 1.16960 1.16960 1.17854 1.18001 1.18001
H2O 76.91887 76.95259 76.96446 76.39888 76.43331 76.44734 76.41974 76.45212 76.46420
F2C=O 314.69819 314.80877 314.83022 312.97864 313.09161 313.11543 313.00795 313.11529 313.13804
HF 100.95377 101.00154 101.01459 100.41148 100.46083 100.47642 100.42746 100.47357 100.48660
CF2: 238.96304 239.04788 239.06437 237.66823 237.75599 237.77466 237.68975 237.77278 237.79055
F∙ 100.20059 100.24050 100.24827 99.70214 99.74344 99.75312 99.71553 99.75374 99.76168
TS3 340.54064 340.66296 340.68668 338.67338 338.79880 338.82528 338.71471 338.83338 338.85836
TS4 416.20303 416.35426 416.38552 —b —b —b 413.95384 414.10040 414.13292
TS5 416.15579 416.30453 416.33567 413.86611 414.01890 414.05357 413.91011 414.05456 414.08767
TS7 339.90747 340.03270 340.05960 338.07929 338.20788 338.23797 338.10715 338.22884 338.25679
TS8 415.60251 415.74883 415.77782 413.35382 413.50350 413.53553 413.39394 413.53538 413.56565
a The energies are given in hartree.
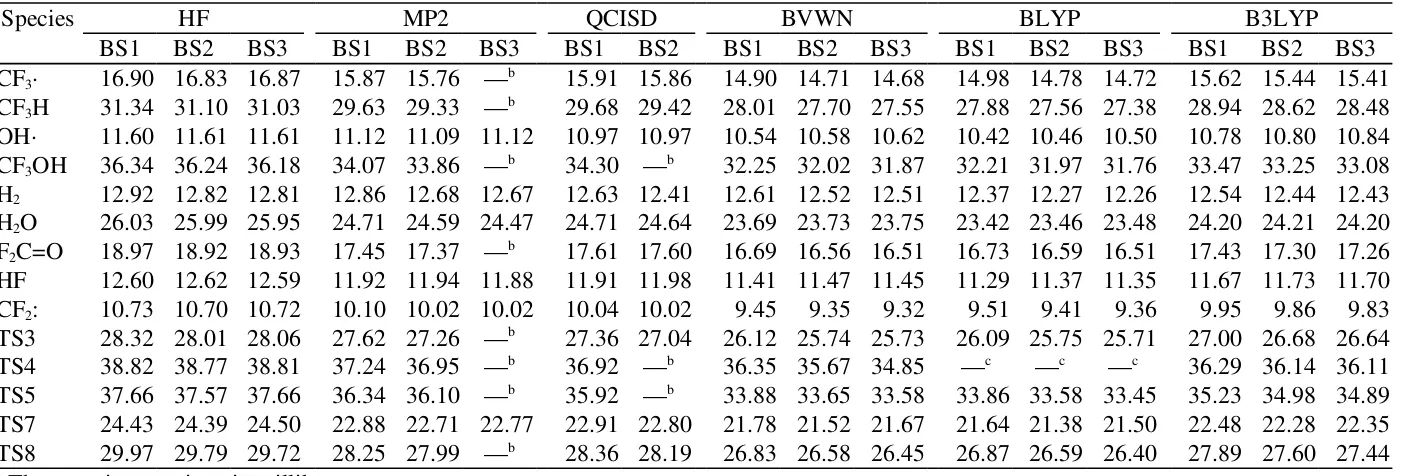
TABLE S3.
The thermal energy corrections at 298.15 K.a
Species HF MP2 QCISD BVWN BLYP B3LYP
BS1 BS2 BS3 BS1 BS2 BS3 BS1 BS2 BS1 BS2 BS3 BS1 BS2 BS3 BS1 BS2 BS3
CF3∙ 16.90 16.83 16.87 15.87 15.76 —b 15.91 15.86 14.90 14.71 14.68 14.98 14.78 14.72 15.62 15.44 15.41
CF3H 31.34 31.10 31.03 29.63 29.33 —b 29.68 29.42 28.01 27.70 27.55 27.88 27.56 27.38 28.94 28.62 28.48
OH∙ 11.60 11.61 11.61 11.12 11.09 11.12 10.97 10.97 10.54 10.58 10.62 10.42 10.46 10.50 10.78 10.80 10.84
CF3OH 36.34 36.24 36.18 34.07 33.86 —b 34.30 —b 32.25 32.02 31.87 32.21 31.97 31.76 33.47 33.25 33.08
H2 12.92 12.82 12.81 12.86 12.68 12.67 12.63 12.41 12.61 12.52 12.51 12.37 12.27 12.26 12.54 12.44 12.43
H2O 26.03 25.99 25.95 24.71 24.59 24.47 24.71 24.64 23.69 23.73 23.75 23.42 23.46 23.48 24.20 24.21 24.20
F2C=O 18.97 18.92 18.93 17.45 17.37 —b 17.61 17.60 16.69 16.56 16.51 16.73 16.59 16.51 17.43 17.30 17.26
HF 12.60 12.62 12.59 11.92 11.94 11.88 11.91 11.98 11.41 11.47 11.45 11.29 11.37 11.35 11.67 11.73 11.70
CF2: 10.73 10.70 10.72 10.10 10.02 10.02 10.04 10.02 9.45 9.35 9.32 9.51 9.41 9.36 9.95 9.86 9.83
TS3 28.32 28.01 28.06 27.62 27.26 —b 27.36 27.04 26.12 25.74 25.73 26.09 25.75 25.71 27.00 26.68 26.64
TS4 38.82 38.77 38.81 37.24 36.95 —b 36.92 —b 36.35 35.67 34.85 —c —c —c 36.29 36.14 36.11
TS5 37.66 37.57 37.66 36.34 36.10 —b 35.92 —b 33.88 33.65 33.58 33.86 33.58 33.45 35.23 34.98 34.89
TS7 24.43 24.39 24.50 22.88 22.71 22.77 22.91 22.80 21.78 21.52 21.67 21.64 21.38 21.50 22.48 22.28 22.35
TS8 29.97 29.79 29.72 28.25 27.99 —b 28.36 28.19 26.83 26.58 26.45 26.87 26.59 26.40 27.89 27.60 27.44
a The energies are given in millihartree.
b Could not calculate.