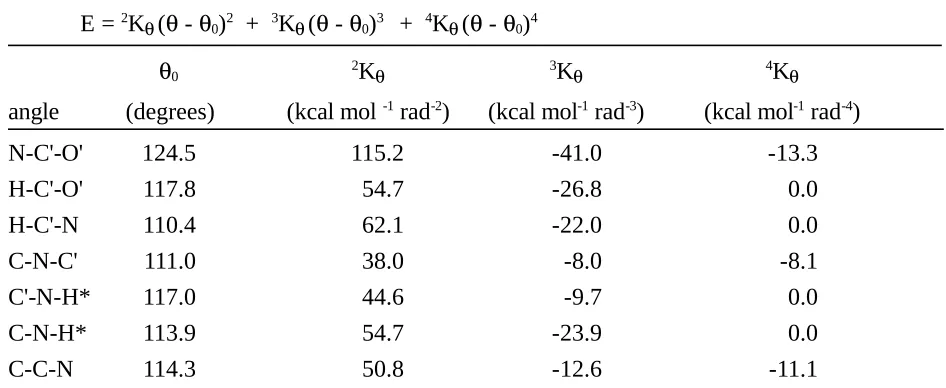
Table I. Force Constants and Reference Values in the QMFF for Amides A. Bond length
E = 2K
b (b - b0)2 + 3Kb(b - b0)3 + 4Kb(b - b0)4
b0 2Kb 3Kb 4Kb
bond (Å) (kcal mol-1 Å-2) (kcal mol-1 Å-3) (kcal mol-1 Å-4)
C'-O' 1.1920
-2568.6 3155.8
C'-N 1.3983 443.4
1415.3
C'-H 1.0934
-780.5 896.3
C-N 1.4416
-676.4 650.0
H*-N 1.0015
-1300.8 1908.3
C-C' 1.5103
-522.3 490.0
H-C 1.0845
-851.3 1040.0
C-C 1.5297
-586.1 758.0
B. Bond angle E = 2K
( - 0)2 + 3K( - 0)3 + 4K( - 0)4
0 2K 3K 4K
angle (degrees) (kcal mol -1 rad-2) (kcal mol-1 rad-3) (kcal mol-1 rad-4)
N-C'-O' 124.5 115.2 -41.0 -13.3
H-C'-O' 117.8 54.7 -26.8 0.0
H-C'-N 110.4 62.1 -22.0 0.0
C-N-C' 111.0 38.0 -8.0 -8.1
C'-N-H* 117.0 44.6 -9.7 0.0
C-N-H* 113.9 54.7 -23.9 0.0
H-C-N 108.9 68.3 3.5 0.0
C-C'-O 123.1 66.1 -20.5 0.2
C-C'-N 116.9 46.9 -13.1 -10.4
H*-N-H* 112.9 46.7 -11.3 0.0
C-C-C' 108.5 61.9 -11.3 -13.1
C'-C-H 107.7 48.3 -34.3 0.0
C-N-C 111.6 47.2 -10.2 -10.1
C'-C-N 100.6 62.0 -6.3 -12.7
N-C'-N 122.5 123.9 -43.7 -28.9
C'-N-C' 122.0 90.8 -31.3 -21.1
H-C-H 107.7 51.5 -8.9 -10.8
H-C-C 110.8 52.7 -10.9 -11.3
C. Torsion angle E = 1K
(1 - cos) + 2K(1 - cos2) + 3K(1 - cos3)
1K
2K 3K
torsion (kcal mol-1) (kcal mol-1) (kcal mol-1)
O'-C'-N-C 0.830 3.723 -0.050
O'-C'-N-H* -1.882 3.043 -0.373
H-C'-N-C 0.334 2.584 -0.401
H-C'-N-H* -0.008 2.619 -0.090
C-C-N-C' 0.020 -0.019 0.013
H-C-N-C' 0.031 -0.037 -0.058
C-C-N-H* -0.069 -0.011 -0.002
H-C-N-H* -0.021 -0.113 -0.175
C-C-C-N 0.108 0.080 -0.287
H-C-C-N -0.025 0.031 -0.207
C-C'-N-H* -0.915 2.385 -0.238
C-C-C'-O' 0.063 0.042 0.080
H-C-C'-O' -0.258 0.002 0.053
C-C-C'-N -0.053 0.056 -0.076
H-C-C'-N 0.241 -0.013 -0.098
C-C-C-C' 0.108 0.080 -0.287
C-C-N-C -0.002 -0.010 0.001
H-C-N-C 0.058 0.051 -0.236
C'-C-C-C' 0.108 0.080 -0.287
C'-C-C-N 0.108 0.080 -0.287
C'-C-N-C' -0.098 0.109 -0.088
C'-C-N-H* 0.078 0.108 0.104
N-C-C'-O' 0.128 0.174 0.129
N-C-C'-N -0.127 0.180 -0.126
C-C'-N-C -0.753 2.739 0.090
C'-C-N-C -0.005 0.007 0.006
N-C'-N-H* -0.818 0.516 -1.233
O'-C'-N-C' -0.452 1.390 -0.834
H-C'-N-C' 0.212 1.246 0.047
H-C-C-H -1.152 0.012 -0.179
D. Out-of-plane E = 2K
2
2K
out-of-plane (kcal mol-1 rad-2)
H-C'-N-O' 42.126
C-N-C'-H* 1.250
C-C'-N-O' 44.242
C'-N-H*-H* 0.000
C-N-C-C' 4.247
N-C'-N-O' 49.203
C'-N-C'-H* 0.000
E. Van der Waals interaction
E = [2(r*/r)9 - 3(r*/r)6] where r* = [(r
i*6 + rj*6) /2] 1/6
and = (ij) 1/2 2 (ri*rj*)3/(ri*6 + rj*6)
ri* i
H* 1.098 0.013
O' 3.535 0.267
C' 3.308 0.120
N 4.070 0.106
H 2.995 0.020
C 4.010 0.054
F. Bond increment
E = 332.0 qi qj / rij; where qi =
k ik. Each ikdonates positive charge to the first atom listedand corresponding negative charge to the second atom. ik
bond (electrons)
C'–O' 0.396
C–N 0.211
H*–N 0.440
C'–N 0.000
H–C' 0.046
C'–C 0.000
H–C 0.053
C–C 0.000
G. Bond/bond
E = Kbb'(b - b0)(b'- b'0) Kbb'
bond/bond (kcal mol-1 Å-2)
O'-C'/C'-N 200.718
O'-C'/C'-H 61.122
N-C'/C'-H 4.058
C'-N/N-C 17.563
C'-N/N-H* -6.250
C-N/N-H* -5.030
N-C/C-C 5.137
N-C/C-H 22.173
C-C'/C'-N -9.389
N-H*/H*-N -0.820
C'-C/C-C 7.855
C'-C/C-H 1.031
N-C/C-N -2.171
N-C/C-C' -5.558
N-C'/C'-N 37.613
H-C/H-C 11.769
H-C/C-C 11.310
H. Bond/angle
E = Kb(b - b0)(- 0)
Kb
bond/angle (kcal mol-1Å-1rad-1)
C'-O'/N-C'-O' 75.9
C'-N/N-C'-O' 90.9
C'-O'/H-C'-O' 71.1
C'-H/H-C'-O' 22.1
C'-N/H-C'-N 45.4
C'-H/H-C'-N 32.4
C'-N/C-N-C' 21.5
C-N/C-N-C' 5.5
C'-N/C'-N-H* 42.9
H*-N/C'-N-H* 15.7
C-N/C-N-H* 17.2
H*-N/C-N-H* 8.6
C-N/C-C-N 6.7
C-C/C-C-N -7.9
C-N/H-C-N 50.6
C-H/H-C-N 15.5
C'-O'/C-C'-O' 53.8
C-C'/C-C'-O' 50.7
C'-N/C-C'-N 8.8
C-C'/C-C'-N 36.8
C-C'/C-C-C' 23.0
C-C/C-C-C' 26.3
C-C'/C'-C-H 18.1
C-H/C'-C-H 13.3
C-N/C-N-C -3.0
C-N/C'-C-N 25.2
C-C'/C'-C-N -7.5
C'-N/N-C'-N 98.7
C'-N/C'-N-C' 29.1
H-C/H-C-H 22.7
H-C/H-C-C 13.9
C-C/H-C-C 34.5
I. Angle/angle
E = K'( - 0)('- '0)
K’
angle/angle common bond (kcal mol-1rad-2)
C-C-N/H-C-N C-N -1.00
C-C-H/C-C-N C-C -4.88
H-C-N/H-C-N C-N 3.71
C-C-H/H-C-N C-H -1.29
H-C-H/H-C-N C-H 6.18
C-C-C'/C'-C-H C-C' 2.96
C-C-C'/C-C-H C-C -2.64
C'-C-H/C'-C-H C-C' -5.07
C-C-H/C'-C-H C-H 1.57
C'-C-H/H-C-H C-H -4.91
C-C-N/C-C-N C-N -0.72
C-C-C/C-C-N C-C 0.22
C'-C-N/H-C-N C-N 4.39
C'-C-H/C'-C-N C-C' 0.08
C'-C-H/H-C-N C-H -2.39
C-C-C'/C'-C-N C-C' 8.24
C-C-C'/C-C-N C-C -1.23
H-C-H/H-C-H H-C 0.93
H-C-H/H-C-C H-C 1.57
H-C-C/H-C-C H-C 3.12
H-C-C/H-C-C C-C -2.18
J. Angle/angle/torsion
E = K’( - 0)(' - '0) cos
K'
angle/angle/torsion (kcal mol-1rad-2)
N-C'-O'/C-N-C'/O'-C'-N-C -22.5
N-C'-O'/C'-N-H*/O'-C'-N-H* -10.6
H-C'-N/C-N-C'/H-C'-N-C -8.1
H-C'-N/C'-N-H*/H-C'-N-H* -7.8
C-C-N/C-N-C'/C-C-N-C' -10.8
H-C-N/C-N-C'/H-C-N-C' -11.8
C-C-N/C-N-H*/C-C-N-H* -6.7
H-C-N/C-N-H*/H-C-N-H* -9.7
C-C-C/C-C-N/C-C-C-N -1.5
C-C-H/C-C-N/H-C-C-N -18.5
C-C'-N/C'-N-H*/C-C'-N-H* -1.9
C-C-C'/C-C'-O'/C-C-C'-O' -11.6
C'-C-H/C-C'-O'/H-C-C'-O' -22.2
C-C-C'/C-C'-N/C-C-C'-N -7.9
C'-C-H/C-C'-N/H-C-C'-N -17.7
C-C-C/C-C-C'/C-C-C-C' -0.6
C-C-C'/C-C-H/C'-C-C-H -7.8
C-C-N/C-N-C/C-C-N-C -2.5
H-C-N/C-N-C/H-C-N-C -17.7
C-C-C'/C-C-C'/C'-C-C-C' 0.2
C-C-C'/C-C-N/C'-C-C-N 0.5
C'-C-N/C-N-C'/C'-C-N-C' -13.4
C'-C-N/C-N-H*/C'-C-N-H* -1.4
C'-C-N/C-C'-O'/N-C-C'-O' -9.5
C-C'-N/C-N-C'/C-C'-N-C -9.5
C'-C-N/C-N-C/C'-C-N-C 0.2
N-C'-N/C'-N-H*/N-C'-N-H* -2.2
N-C'-O'/C'-N-C'/O'-C'-N-C' -4.9
H-C'-N/C'-N-C'/H-C'-N-C' -1.1
H-C-C/H-C-C/H-C-C-H -14.8
K. Bond/torsion(Type 1)
E = (b- b0) [1Kbcos + 2Kbcos2 + 3Kbcos3
1K
b 2Kb 3Kb
bond/torsion (kcal mol-1Å-1) (kcal mol-1Å-1) (kcal mol-1Å-1)
C'-N/O'-C'-N-C -12.797 20.736 -2.568
C'-N/O'-C'-N-H* -1.316 8.905 -0.703
C'-N/H-C'-N-C -16.993 4.645 -4.365
C'-N/H-C'-N-H* -1.399 7.542 -3.346
C-N/C-C-N-C' -5.725 -0.580 -0.985
C-N/H-C-N-C' -0.100 -3.282 1.678
C-N/C-C-N-H* -5.131 -4.908 0.051
C-N/H-C-N-H* -1.703 4.066 1.171
C-C/C-C-C-N -6.134 -4.786 -1.919
C-C/H-C-C-N -5.946 -0.861 -0.068
C'-N/C-C'-N-H* -0.768 6.863 -1.542
C-C'/C-C-C'-O' 0.491 -0.159 0.177
C-C'/H-C-C'-O' 0.342 1.324 1.390
C-C'/C-C-C'-N -2.907 -2.236 2.767
C-C'/H-C-C'-N 0.333 -0.601 1.160
C-C/C-C-C-C' -2.311 0.328 -1.002
C-C/C'-C-C-H -5.078 1.806 -1.103
C-N/C-C-N-C -6.822 -1.530 -4.325
C-N/H-C-N-C -3.323 0.472 1.400
C-C/C'-C-C-C' 2.139 0.848 -0.270
C-C/C'-C-C-N -1.770 -5.899 -4.924
C-N/C'-C-N-C' -6.875 -7.398 -7.869
C-N/C'-C-N-H* -2.104 5.859 -0.765
C-C'/N-C-C'-O' -6.527 7.099 0.619
C'-N/C-C'-N-C -13.408 4.941 -4.111
C-N/C'-C-N-C 1.828 -5.164 -4.479
C'-N/N-C'-N-H* -1.803 -6.429 3.201
C'-N/O'-C'-N-C' -0.162 -1.738 0.983
C'-N/H-C'-N-C' -0.699 -0.856 -1.197
C-C/H-C-C-H -16.582 -0.784 -0.901
L. Bond/torsion(type 2)
E =(b'- b'0) [1Kb' cos + 2Kb'cos2 + 3Kb'cos3
1K
b' 2Kb' 3Kb'
bond/torsion (kcal mol-1Å-1) (kcal mol-1Å-1) (kcal mol-1Å-1)
C'-O'/O'-C'-N-C 0.178 -3.091 0.809
C-N/O'-C'-N-C 0.232 1.051 -0.146
C'-O'/O'-C'-N-H* -1.102 -3.831 1.807
H*-N/O'-C'-N-H 0.176 0.281 0.118
C'-H/H-C'-N-C -0.859 -0.007 0.215
C-N/H-C'-N-C -0.773 0.830 -0.094
C'-H/H-C'-N-H* -0.372 0.693 0.517
H*-N/H-C'-N-H* 0.143 0.808 0.170
C'-N/C-C-N-C' -0.180 -1.358 1.128
C-C/C-C-N-C' -0.295 0.005 0.081
C'-N/H-C-N-C' 0.332 1.700 -0.084
C-H/H-C-N-C' -0.531 1.188 0.194
H*-N/C-C-N-H* -0.144 -0.105 -0.600
C-C/C-C-N-H* 0.191 0.002 0.192
H*-N/H-C-N-H* -0.709 0.238 0.450
C-H/H-C-N-H* -1.302 0.410 0.128
C-N/C-C-C-N -0.115 -0.059 0.037
C-C/C-C-C-N 0.108 0.015 0.075
C-N/H-C-C-N 0.438 0.364 0.673
C-H/H-C-C-N -0.087 -0.545 -0.272
C-C'/C-C'-N-H* -1.011 0.814 0.610
H*-N/C-C'-N-H* 0.929 0.243 0.209
C'-O'/C-C-C'-O' -0.407 0.118 -0.221
C-C/C-C-C'-O' 0.385 0.073 0.152
C-H/H-C-C'-O' 1.760 0.410 0.568
C'-N/C-C-C'-N -0.399 -0.443 -0.256
C-C/C-C-C'-N -0.381 -0.011 -0.166
C'-N/H-C-C'-N 0.515 -0.389 0.845
C-H/H-C-C'-N -0.039 1.136 0.005
C-C'/C-C-C-C' 0.008 0.009 -0.001
C-C/C-C-C-C' 0.009 0.000 0.005
C-C'/C'-C-C-H -0.030 0.526 -0.642
C-H/C'-C-C-H -0.014 -0.046 -0.109
C-N/C-C-N-C -0.119 -0.011 -0.135
C-C/C-C-N-C -0.181 -0.164 -0.112
C-N/H-C-N-C 0.181 0.470 -0.108
C-H/H-C-N-C -1.954 1.159 0.979
C-C'/C'-C-C-C' 0.008 -0.001 -0.006
C-N/C'-C-C-N -0.244 -0.214 -0.214
C-C'/C'-C-C-N -0.191 -0.161 -0.168
C'-N/C'-C-N-C' -0.645 0.617 -0.631
C-C'/C'-C-N-C' -0.259 0.311 -0.406
H*-N/C'-C-N-H* 1.305 0.430 0.085
C-C'/C'-C-N-H* 0.158 0.487 0.134
C-N/N-C-C'-O' -0.110 0.379 0.234
C'-O'/N-C-C'-O' 1.456 -0.479 0.945
C-N/N-C-C'-N -0.252 0.104 -0.865
C'-N/N-C-C'-N -0.018 0.028 -1.389
C-C'/C-C'-N-C 0.135 -0.671 0.418
C-N/C-C'-N-C 0.333 -0.165 -0.206
C-N/C'-C-N-C 0.026 -0.131 -0.117
C-C'/C'-C-N-C 0.097 -0.115 -0.134
H*-N/N-C'-N-H* -0.707 -1.033 0.191
C'-N/N-C'-N-H* 0.112 -0.767 -0.006
C'-O'/O'-C'-N-C' -1.017 1.204 -0.996
C'-N/O'-C'-N-C' 0.250 -0.699 0.386
C'-N/H-C'-N-C' -0.306 -0.609 -0.330
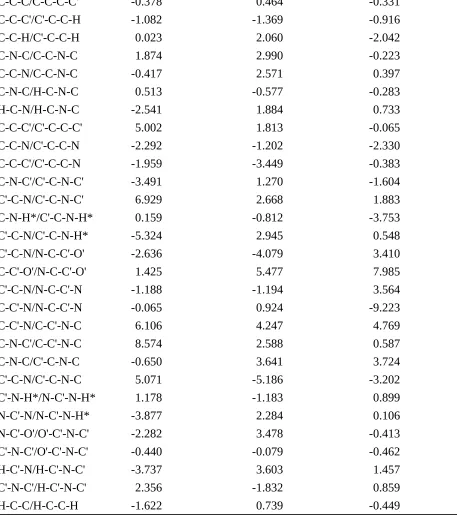
M. Angle/torsion
E = (- 0) [1Kcos + 2Kcos2 + 3Kcos3 1K
2K 3K
angle/torsion (kcal mol-1rad-1) (kcal mol-1rad-1) (kcal mol-1rad-1)
N-C'-O'/O'-C'-N-C 6.444 5.843 2.482
C-N-C'/O'-C'-N-C 10.787 3.117 -0.320
N-C'-O'/O'-C'-N-H* -3.803 0.523 0.793
C'-N-H*/O'-C'-N-H* 6.115 1.803 0.359
H-C'-N/H-C'-N-C 0.197 4.539 0.310
C-N-C'/H-C'-N-C 8.960 -0.511 -0.311
H-C'-N/H-C'-N-H* -3.163 2.963 -0.211
C'-N-H*/H-C'-N-H* 5.694 1.463 -0.046
C-N-C'/C-C-N-C' -2.207 1.637 1.039
C-C-N/C-C-N-C' -1.095 0.082 1.765
C-N-C'/H-C-N-C' 0.054 -0.495 -0.112
H-C-N/H-C-N-C' -2.197 3.012 0.777
C-N-H*/C-C-N-H* -1.489 0.523 -0.355
C-C-N/C-C-N-H* -4.198 3.925 -0.054
C-N-H*/H-C-N-H* -0.992 0.523 0.114
H-C-N/H-C-N-H* -5.365 2.011 0.347
C-C-N/C-C-C-N 0.295 0.232 -1.152
C-C-C/C-C-C-N -0.797 -2.461 0.360
C-C-N/H-C-C-N -2.872 0.337 -0.569
C-C-H/H-C-C-N -1.807 2.454 -1.751
C-C'-N/C-C'-N-H* -3.208 1.871 1.410
C'-N-H*/C-C'-N-H* 4.950 0.540 0.143
C-C'-O'/C-C-C'-O' 0.978 0.864 0.975
C-C-C'/C-C-C'-O' 0.128 -1.986 -0.790
C-C'-O'/H-C-C'-O' -2.166 1.059 -0.302
C'-C-H/H-C-C'-O' 13.232 -0.703 0.519
C-C'-N/C-C-C'-N 3.076 0.729 -0.111
C-C-C'/C-C-C'-N 3.160 -0.049 -2.002
C-C'-N/H-C-C'-N 2.900 0.735 1.218
C'-C-H/H-C-C'-N 10.283 0.011 1.001
C-C-C/C-C-C-C' -0.378 0.464 -0.331
C-C-C'/C'-C-C-H -1.082 -1.369 -0.916
C-C-H/C'-C-C-H 0.023 2.060 -2.042
C-N-C/C-C-N-C 1.874 2.990 -0.223
C-C-N/C-C-N-C -0.417 2.571 0.397
C-N-C/H-C-N-C 0.513 -0.577 -0.283
H-C-N/H-C-N-C -2.541 1.884 0.733
C-C-C'/C'-C-C-C' 5.002 1.813 -0.065
C-C-N/C'-C-C-N -2.292 -1.202 -2.330
C-C-C'/C'-C-C-N -1.959 -3.449 -0.383
C-N-C'/C'-C-N-C' -3.491 1.270 -1.604
C'-C-N/C'-C-N-C' 6.929 2.668 1.883
C-N-H*/C'-C-N-H* 0.159 -0.812 -3.753
C'-C-N/C'-C-N-H* -5.324 2.945 0.548
C'-C-N/N-C-C'-O' -2.636 -4.079 3.410
C-C'-O'/N-C-C'-O' 1.425 5.477 7.985
C'-C-N/N-C-C'-N -1.188 -1.194 3.564
C-C'-N/N-C-C'-N -0.065 0.924 -9.223
C-C'-N/C-C'-N-C 6.106 4.247 4.769
C-N-C'/C-C'-N-C 8.574 2.588 0.587
C-N-C/C'-C-N-C -0.650 3.641 3.724
C'-C-N/C'-C-N-C 5.071 -5.186 -3.202
C'-N-H*/N-C'-N-H* 1.178 -1.183 0.899
N-C'-N/N-C'-N-H* -3.877 2.284 0.106
N-C'-O'/O'-C'-N-C' -2.282 3.478 -0.413
C'-N-C'/O'-C'-N-C' -0.440 -0.079 -0.462
H-C'-N/H-C'-N-C' -3.737 3.603 1.457
C'-N-C'/H-C'-N-C' 2.356 -1.832 0.859
H-C-C/H-C-C-H -1.622 0.739 -0.449
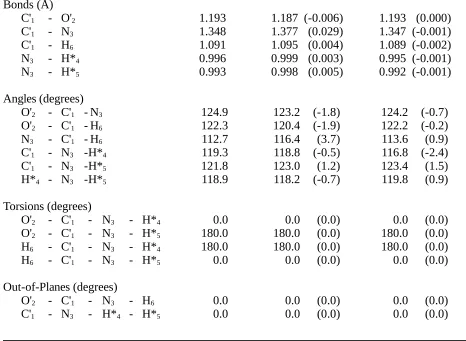
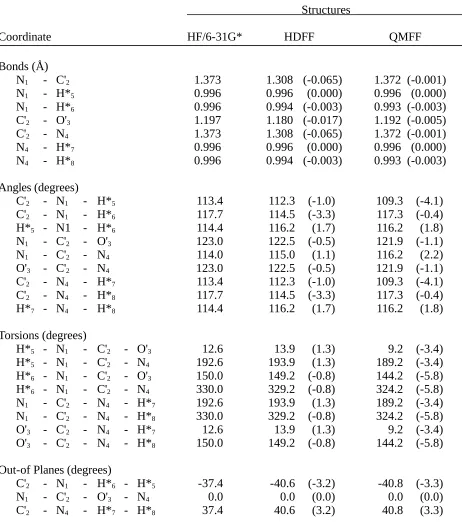
Table II.A. Comparison of Formamide Structures Optimizedby Ab Initio
Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Bonds (Å)
C'1 - O'2 1.193 1.187 (-0.006) 1.193 (0.000)
C'1 - N3 1.348 1.377 (0.029) 1.347 (-0.001)
C'1 - H6 1.091 1.095 (0.004) 1.089 (-0.002)
N3 - H*4 0.996 0.999 (0.003) 0.995 (-0.001)
N3 - H*5 0.993 0.998 (0.005) 0.992 (-0.001)
Angles (degrees)
O'2 - C'1 - N3 124.9 123.2 (-1.8) 124.2 (-0.7)
O'2 - C'1 - H6 122.3 120.4 (-1.9) 122.2 (-0.2)
N3 - C'1 - H6 112.7 116.4 (3.7) 113.6 (0.9)
C'1 - N3 -H*4 119.3 118.8 (-0.5) 116.8 (-2.4)
C'1 - N3 -H*5 121.8 123.0 (1.2) 123.4 (1.5)
H*4 - N3 -H*5 118.9 118.2 (-0.7) 119.8 (0.9)
Torsions (degrees)
O'2 - C'1 - N3 - H*4 0.0 0.0 (0.0) 0.0 (0.0)
O'2 - C'1 - N3 - H*5 180.0 180.0 (0.0) 180.0 (0.0)
H6 - C'1 - N3 - H*4 180.0 180.0 (0.0) 180.0 (0.0)
H6 - C'1 - N3 - H*5 0.0 0.0 (0.0) 0.0 (0.0)
Out-of-Planes (degrees)
O'2 - C'1 - N3 - H6 0.0 0.0 (0.0) 0.0 (0.0)
C'1 - N3 - H*4 - H*5 0.0 0.0 (0.0) 0.0 (0.0)
Table II.B. Comparison of Planar Acetamide Structures Optimizedby Ab Initio
Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
Bonds (Å)
O'1 - C'2 1.198 1.187 (-0.011) 1.196 (-0.002)
C'2 - N3 1.356 1.369 (0.013) 1.352 (-0.004)
C'2 - C4 1.514 1.509 (-0.005) 1.503 (-0.011)
N3 - H*5 0.995 0.998 (0.003) 0.995 (0.000)
N3 - H*6 0.993 0.996 (0.003) 0.990 (-0.002)
C4 - H7 1.085 1.085 (0.000) 1.084 (-0.001)
C4 - H8 1.085 1.085 (0.000) 1.084 (-0.001)
C4 - H9 1.080 1.085 (0.005) 1.079 (-0.001)
O'1 - C'2 - N3 122.2 121.3 (-0.9) 121.4 (-0.8)
O'1 - C'2 - C4 122.9 123.4 (0.5) 123.0 (0.1)
N3 - C'2 - C4 114.9 115.3 (0.4) 115.7 (0.7)
C'2 - N3 -H*5 118.5 118.8 (0.3) 116.5 (-2.1)
C'2 - N3 -H*6 122.7 122.6 (-0.1) 123.5 (0.8)
H*5 - N3 -H*6 118.7 118.6 (-0.1) 120.1 (1.3)
C'2 - C4 - H7 110.6 109.4 (-1.2) 110.7 (0.1)
C'2 - C4 - H8 110.6 109.4 (-1.2) 110.7 (0.1)
C'2 - C4 - H9 108.9 111.2 (2.3) 109.3 (0.4)
H7 - C4 - H8 108.0 106.9 (-1.1) 108.9 (0.9)
H7 - C4 - H9 109.4 109.9 (0.5) 108.6 (-0.8)
H8 - C4 - H9 109.4 109.9 (0.5) 108.6 (-0.8)
Torsions (degrees)
O'1 - C'2 - N3 - H*5 0.0 0.0 (0.0) 0.0 (0.0)
O'1 - C'2 - N3 - H*6 180.0 180.0 (0.0) 180.0 (0.0)
C4 - C'2 - N3 - H*5 180.0 180.0 (0.0) 180.0 (0.0)
C4 - C'2 - N3 - H*6 0.0 0.0 (0.0) 0.0 (0.0)
O'1 - C'2 - C4 - H7 120.2 121.6 (1.4) 119.6 (-0.6)
O'1 - C'2 - C4 - H8 239.8 238.4 (-1.4) 240.4 (0.6)
O'1 - C'2 - C4 - H9 0.0 0.0 (0.0) 0.0 (0.0)
N3 - C'2 - C4 - H7 300.2 301.6 (1.4) 299.6 (-0.6)
N3 - C'2 - C4 - H8 59.8 58.4 (-1.4) 60.4 (0.6)
N3 - C'2 - C4 - H9 180.0 180.0 (0.0) 180.0 (0.0)
Out-of-Planes (degrees)
O'1 - C'2 - C4 - N3 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - H*6 - H*5 0.0 0.0 (0.0) 0.0 (0.0)
Appendix II.C. Comparison of Planar N-Methylformamide Trans and Cis Structures Optimized by Ab Initio Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
trans cis
Coordinate HF/6-31G* HDFF QMFF HF/6-31G* HDFF QMFF
Bonds (Å)
O'1 -C'2 1.196 1.190 (-0.006) 1.196 (0.001) 1.195 1.189 (-0.006) 1.194 (-0.001)
C'2 -N3 1.345 1.401 (0.056) 1.348 (0.003) 1.347 1.389 (0.041) 1.349 (0.002)
C'2 -H5 1.091 1.094 (0.003) 1.088 (-0.003) 1.091 1.095 (0.004) 1.092 (0.000)
N3 - C4 1.448 1.452 (0.004) 1.445 (-0.003) 1.442 1.450 (0.007) 1.440 (-0.002)
N3 -H*6 0.993 0.999 (0.006) 0.995 (0.002) 0.997 1.001 (0.004) 1.000 (0.003)
C4 -H7 1.081 1.085 (0.004) 1.083 (0.002) 1.082 1.086 (0.003) 1.082 (0.000)
C4 -H8 1.083 1.086 (0.003) 1.085 (0.002) 1.084 1.086 (0.001) 1.085 (0.001)
C4 -H9 1.083 1.086 (0.003) 1.085 (0.002) 1.084 1.086 (0.001) 1.085 (0.001)
Angles (degrees)
O'1 -C'2 -H5 122.2 119.6 (-2.7) 121.8 (-0.4) 122.1 120.0 (-2.2) 121.9 (-0.2)
N3 -C'2 -H5 113.0 114.6 (1.6) 113.6 (0.6) 112.9 116.6 (3.7) 113.0 (0.0)
C'2 -N3 - C4 121.8 122.5 (0.7) 120.8 (-0.9) 125.4 124.5 (-0.9) 124.5 (-0.9)
C'2 -N3 -H*6 118.7 120.0 (1.3) 120.7 (2.0) 115.5 117.1 (1.6) 115.4 (-0.1)
C4 -N3 -H*6 119.6 117.5 (-2.0) 118.5 (-1.1) 119.1 118.4 (-0.7) 120.1 (1.0)
N3 - C4 -H7 108.7 110.2 (1.5) 109.0 (0.3) 109.3 110.7 (1.5) 108.3 (-1.0)
N3 - C4 -H8 110.9 110.7 (-0.2) 111.1 (0.2) 111.4 110.5 (-0.8) 111.4 (0.0)
N3 - C4 -H9 110.9 110.7 (-0.2) 111.1 (0.2) 111.3 110.5 (-0.8) 111.4 (0.1)
H7 - C4 -H8 109.0 107.9 (-1.1) 108.4 (-0.5) 108.1 107.9 (-0.2) 108.5 (0.4)
H7 - C4 -H9 109.0 107.9 (-1.1) 108.4 (-0.5) 108.1 107.9 (-0.1) 108.5 (0.4)
H8 - C4 -H9 108.4 109.3 (1.0) 108.7 (0.3) 108.7 109.1 (0.5) 108.7 (0.0)
Torsions (degrees)
O'1 -C'2 -N3 - C4 0.0 0.0 (0.0) 0.0 (0.0) 180.1 180.0 (-0.1) 180.0 (-0.1)
O'1 -C'2 -N3 -H*6 180.0 180.0 (0.0) 180.0 (0.0) 359.9 0.0 (0.1) 0.0 (0.1)
H5 -C'2 -N3 - C4 180.0 180.0 (0.0) 180.0 (0.0) 0.1 0.0 (-0.1) 0.0 (-0.1)
H5 -C'2 -N3 -H*6 0.0 0.0 (0.0) 0.0 (0.0) 179.9 180.0 (0.1) 180.0 (0.1)
C'2 -N3 - C4 -H7 180.0 180.0 (0.0) 180.0 (0.0) 359.7 0.0 (0.3) 0.0 (0.3)
C'2 -N3 - C4 -H8 299.8 299.3 (-0.5) 299.4 (-0.3) 119.0 119.5 (0.6) 119.2 (0.3)
C'2 -N3 - C4 -H9 60.2 60.7 (0.5) 60.6 (0.3) 240.4 240.5 (0.1) 240.8 (0.4)
H*6 -N3 - C4 -H7 0.0 0.0 (0.0) 0.0 (0.0) 179.9 180.0 (0.1) 180.0 (0.1)
H*6 -N3 - C4 -H8 119.8 119.3 (-0.5) 119.4 (-0.3) 299.2 299.5 (0.4) 299.2 (0.1)
H*6 -N3 - C4 -H9 240.2 240.7 (0.5) 240.6 (0.3) 60.6 60.5 (-0.2) 60.8 (0.1)
Out-of-Planes (degrees) O’
'1-C'2 -N3 -H5 0.0
(0.0)0.0(0.0)0.00.0 (0.0 0.0(0.0)
C'2 -N3 -H*6 - C4 0.0 (0.0)0.0(0.0)0.00.0 (0.0 0.0(0.0)
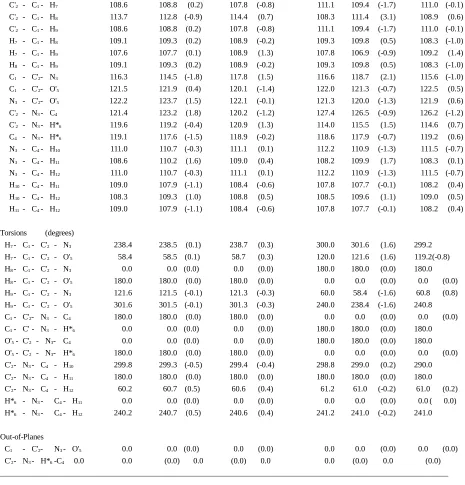
Appendix II.D. Comparison of Planar N-Methylacetamide Trans and Cis Structures Optimized by Ab Initio Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
trans cis
Coordinate HF/6-31G* HDFF QMFF HF/6-31G* HDFF QMFF
Bonds (Å)
Angles (degrees)
C'2 - C1- H7 108.6 108.8 (0.2) 107.8 (-0.8) 111.1 109.4 (-1.7) 111.0 (-0.1) C'2 - C1- H8 113.7 112.8 (-0.9) 114.4 (0.7) 108.3 111.4 (3.1) 108.9 (0.6) C'2 - C1- H9 108.6 108.8 (0.2) 107.8 (-0.8) 111.1 109.4 (-1.7) 111.0 (-0.1) H7 - C1- H8 109.1 109.3 (0.2) 108.9 (-0.2) 109.3 109.8 (0.5) 108.3 (-1.0) H7 - C1- H9 107.6 107.7 (0.1) 108.9 (1.3) 107.8 106.9 (-0.9) 109.2 (1.4) H8 - C1- H9 109.1 109.3 (0.2) 108.9 (-0.2) 109.3 109.8 (0.5) 108.3 (-1.0) C1 - C'2- N3 116.3 114.5 (-1.8) 117.8 (1.5) 116.6 118.7 (2.1) 115.6 (-1.0) C1 - C'2- O'5 121.5 121.9 (0.4) 120.1 (-1.4) 122.0 121.3 (-0.7) 122.5 (0.5) N3 - C'2- O'5 122.2 123.7 (1.5) 122.1 (-0.1) 121.3 120.0 (-1.3) 121.9 (0.6) C'2 - N3- C4 121.4 123.2 (1.8) 120.2 (-1.2) 127.4 126.5 (-0.9) 126.2 (-1.2) C'2 - N3- H*6 119.6 119.2 (-0.4) 120.9 (1.3) 114.0 115.5 (1.5) 114.6 (0.7) C4 - N3- H*6 119.1 117.6 (-1.5) 118.9 (-0.2) 118.6 117.9 (-0.7) 119.2 (0.6) N3 - C4- H10 111.0 110.7 (-0.3) 111.1 (0.1) 112.2 110.9 (-1.3) 111.5 (-0.7) N3 - C4- H11 108.6 110.2 (1.6) 109.0 (0.4) 108.2 109.9 (1.7) 108.3 (0.1) N3 - C4- H12 111.0 110.7 (-0.3) 111.1 (0.1) 112.2 110.9 (-1.3) 111.5 (-0.7) H10- C4- H11 109.0 107.9 (-1.1) 108.4 (-0.6) 107.8 107.7 (-0.1) 108.2 (0.4) H10- C4- H12 108.3 109.3 (1.0) 108.8 (0.5) 108.5 109.6 (1.1) 109.0 (0.5) H11 - C4- H12 109.0 107.9 (-1.1) 108.4 (-0.6) 107.8 107.7 (-0.1) 108.2 (0.4)
Torsions (degrees)
H7- C1- C'2 - N3 238.4 238.5 (0.1) 238.7 (0.3) 300.0 301.6 (1.6) 299.2 H7- C1- C'2 - O'5 58.4 58.5 (0.1) 58.7 (0.3) 120.0 121.6 (1.6) 119.2(-0.8) H8- C1- C'2 - N3 0.0 0.0 (0.0) 0.0 (0.0) 180.0 180.0 (0.0) 180.0 H8- C1- C'2 - O'5 180.0 180.0 (0.0) 180.0 (0.0) 0.0 0.0 (0.0) 0.0 (0.0) H9- C1- C'2 - N3 121.6 121.5 (-0.1) 121.3 (-0.3) 60.0 58.4 (-1.6) 60.8 (0.8) H9- C1- C'2 - O'5 301.6 301.5 (-0.1) 301.3 (-0.3) 240.0 238.4 (-1.6) 240.8 C1- C'2- N3 - C4 180.0 180.0 (0.0) 180.0 (0.0) 0.0 0.0 (0.0) 0.0 (0.0) C1- C' - N3 - H*6 0.0 0.0 (0.0) 0.0 (0.0) 180.0 180.0 (0.0) 180.0 O'5- C'2 - N3- C4 0.0 0.0 (0.0) 0.0 (0.0) 180.0 180.0 (0.0) 180.0 O'5- C'2 - N3- H*6 180.0 180.0 (0.0) 180.0 (0.0) 0.0 0.0 (0.0) 0.0 (0.0) C'2- N3- C4 - H10 299.8 299.3 (-0.5) 299.4 (-0.4) 298.8 299.0 (0.2) 290.0 C'2- N3- C4 - H11 180.0 180.0 (0.0) 180.0 (0.0) 180.0 180.0 (0.0) 180.0 C'2- N3- C4 - H12 60.2 60.7 (0.5) 60.6 (0.4) 61.2 61.0 (-0.2) 61.0 (0.2) H*6 - N3- C4- H11 0.0 0.0 (0.0) 0.0 (0.0) 0.0 0.0 (0.0) 0.0 ( 0.0) H*6 - N3- C4- H12 240.2 240.7 (0.5) 240.6 (0.4) 241.2 241.0 (-0.2) 241.0
Out-of-Planes
C1 - C'2- N3- O'5 0.0 0.0 (0.0) 0.0 (0.0) 0.0 0.0 (0.0) 0.0 (0.0) C'2- N3- H*6-C4 0.0 0.0 (0.0) 0.0 (0.0) 0.0 0.0 (0.0) 0.0 (0.0) ______________________________________________________________________________________________________________
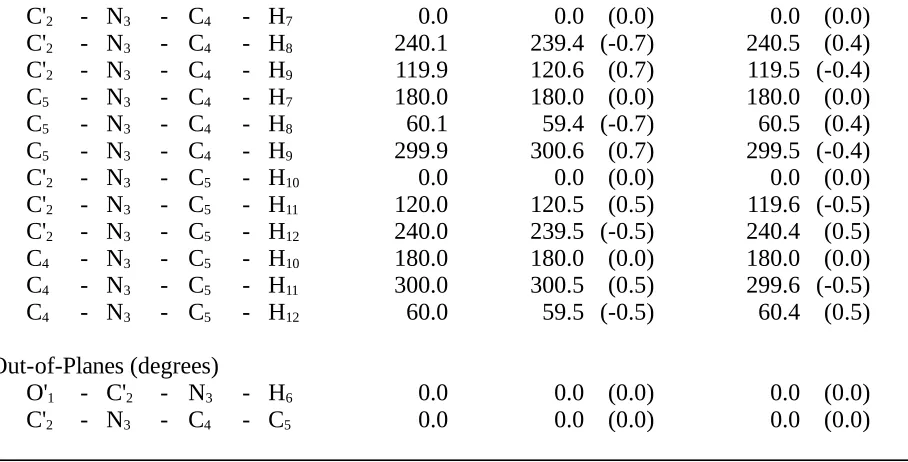
Table II.E. Comparison of N,N-Dimethylformamide Structures Optimizedby Ab Initio
Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
O'1 - C'2 1.196 1.192 (-0.004) 1.197 (0.001)
C'2 - N3 1.349 1.415 (0.067) 1.350
(0.002)
C'2 - H6 1.091 1.094 (0.004) 1.091
(0.001)
N3 - C4 1.442 1.454 (0.012) 1.447
(0.005)
N3 - C5 1.446 1.457 (0.011) 1.448
(0.002)
C4 - H7 1.082 1.085 (0.003) 1.080
(-0.002)
C4 - H8 1.086 1.086 (0.000) 1.087
(0.001)
C4 - H9 1.086 1.086 (0.000) 1.087
(0.001)
C5 - H10 1.077 1.085 (0.008) 1.080
(0.003)
C5 - H11 1.086 1.085 (-0.001) 1.087
(0.001)
C5 - H12 1.086 1.085 (-0.001) 1.087
(0.001)
Angles (degrees)
O'1 - C'2 - N3 125.9 126.1 (0.2) 126.2 (0.3)
O'1 - C'2 - H6 121.5 118.8 (-2.7) 120.9 (-0.6)
N3 - C'2 - H6 112.6 115.0 (2.5) 112.9 (0.3)
C'2 - N3 - C4 122.0 121.3 (-0.7) 121.4 (-0.6)
C'2 - N3 - C5 120.6 122.9 (2.3) 122.1 (1.5)
C4 - N3 - C5 117.4 115.8 (-1.6) 116.6 (-0.9)
N3 - C4 - H7 110.2 111.4 (1.3) 109.3 (-0.9)
N3 - C4 - H8 110.9 110.4 (-0.5) 111.1 (0.2)
N3 - C4 - H9 110.9 110.4 (-0.5) 111.1 (0.2)
H7 - C4 - H8 108.3 108.4 (0.1) 108.3 (0.1)
H7 - C4 - H9 108.3 108.4 (0.1) 108.3 (0.1)
H8 - C4 - H9 108.2 107.6 (-0.6) 108.6 (0.3)
N3 - C5 - H10 109.0 111.7 (2.8) 109.9 (0.9)
N3 - C5 - H11 110.5 110.4 (-0.1) 111.0 (0.6)
N3 - C5 - H12 110.5 110.4 (-0.1) 111.0 (0.6)
H10 - C5 - H11 109.3 108.3 (-1.0) 108.1 (-1.1)
H10 - C5 - H12 109.3 108.3 (-1.0) 108.1 (-1.1)
H11 - C5 - H12 108.4 107.7 (-0.7) 108.5 (0.1)
Torsions (degrees)
O'1 - C'2 - N3 - C4 180.0 180.0 (0.0) 180.0 (0.0)
O'1 - C'2 - N3 - C5 0.0 0.0 (0.0) 0.0 (0.0)
H6 - C'2 - N3 - C4 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - C4 - H7 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - C4 - H8 240.1 239.4 (-0.7) 240.5 (0.4)
C'2 - N3 - C4 - H9 119.9 120.6 (0.7) 119.5 (-0.4)
C5 - N3 - C4 - H7 180.0 180.0 (0.0) 180.0 (0.0)
C5 - N3 - C4 - H8 60.1 59.4 (-0.7) 60.5 (0.4)
C5 - N3 - C4 - H9 299.9 300.6 (0.7) 299.5 (-0.4)
C'2 - N3 - C5 - H10 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - C5 - H11 120.0 120.5 (0.5) 119.6 (-0.5)
C'2 - N3 - C5 - H12 240.0 239.5 (-0.5) 240.4 (0.5)
C4 - N3 - C5 - H10 180.0 180.0 (0.0) 180.0 (0.0)
C4 - N3 - C5 - H11 300.0 300.5 (0.5) 299.6 (-0.5)
C4 - N3 - C5 - H12 60.0 59.5 (-0.5) 60.4 (0.5)
Out-of-Planes (degrees)
O'1 - C'2 - N3 - H6 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - C4 - C5 0.0 0.0 (0.0) 0.0 (0.0)
Table II.F. Comparison of Planar N,N-Dimethylacetamide Structures Optimized by Ab Initio
Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
Bonds (Å)
C1 - C'2 1.518 1.520 (0.002) 1.521 (0.004)
C1 - H7 1.079 1.085 (0.006) 1.080 (0.001)
C1 - H8 1.085 1.084 (0.000) 1.084 (-0.001)
C1 - H9 1.085 1.084 (0.000) 1.084 (-0.001)
C'2 - O'3 1.203 1.192 (-0.010) 1.201 (-0.001)
C'2 - N4 1.361 1.430 (0.069) 1.366 (0.006)
N4 - C5 1.449 1.461 (0.012) 1.455 (0.006)
N4 - C6 1.445 1.459 (0.014) 1.450 (0.005)
C5 - H10 1.076 1.085 (0.009) 1.079 (0.004)
C5 - H11 1.087 1.085 (-0.001) 1.087 (0.001)
C5 - H12 1.087 1.085 (-0.001) 1.087 (0.001)
C6 - H13 1.077 1.083 (0.006) 1.078 (0.001)
C6 - H14 1.087 1.086 (-0.001) 1.088 (0.001)
C6 - H15 1.087 1.086 (-0.001) 1.088 (0.001)
Angles (degrees)
C'2 - C1 - H7 107.2 111.4 (4.2) 108.1 (0.9)
C'2 - C1 - H8 111.6 109.5 (-2.1) 111.4 (-0.3)
H7 - C1 - H8 109.1 109.5 (0.5) 108.3 (-0.8)
H7 - C1 - H9 109.1 109.5 (0.5) 108.3 (-0.8)
H8 - C1 - H9 108.1 107.3 (-0.9) 109.3 (1.1)
C1 - C'2 - O'3 119.9 119.0 (-0.9) 119.0 (-0.8)
C1 - C'2 - N4 118.0 118.3 (0.3) 118.8 (0.8)
O'3 - C'2 - N4 122.1 122.1 (0.6) 122.2 (0.1)
C'2 - N4 - C5 119.5 121.4 (1.8) 120.9 (1.4)
C'2 - N4 - C6 125.6 124.8 (-0.8) 124.9 (-0.7)
C5 - N4 - C6 114.9 113.9 (-1.0) 114.2 (-0.6)
N4 - C5 - H10 109.5 112.4 (3.0) 110.8 (1.4)
N4 - C5 - H11 110.3 110.3 (0.0) 110.8 (0.4)
N4 - C5 - H12 110.3 110.3 (0.0) 110.8 (0.4)
H10 - C5 - H11 109.3 108.0 (-1.2) 107.9 (-1.3)
H10 - C5 - H12 109.3 108.0 (-1.2) 107.9 (-1.3)
H11 - C5 - H12 108.2 107.5 (-0.6) 108.5 (0.3)
N4 - C6 - H13 111.8 113.0 (1.1) 111.1 (-0.7)
N4 - C6 - H14 110.4 110.2 (-0.2) 110.6 (0.3)
N4 - C6 - H15 110.4 110.2 (-0.2) 110.6 (0.3)
H13 - C6 - H14 108.1 108.0 (-0.1) 108.0 (-0.1)
H13 - C6 - H15 108.1 108.0 (-0.1) 108.0 (-0.1)
H14 - C6 - H15 107.9 107.4 (-0.6) 108.4 (0.5)
Torsions (degrees)
H7 - C1 - C'2 - O'3 0.0 0.0 (0.0) 0.0 (0.0)
H7 - C1 - C'2 - N4 180.0 180.0 (0.0) 180.0 (0.0)
H8 - C1 - C'2 - O'3 119.4 121.3 (1.9) 118.9 (-0.6)
H8 - C1 - C'2 - N4 299.4 301.3 (1.9) 298.9 (-0.6)
H9 - C1 - C'2 - O'3 240.6 238.7 (-1.9) 241.1 (0.6)
H9 - C1 - C'2 - N4 60.6 58.7 (-1.9) 61.1 (0.6)
C1 - C'2 - N4 - C5 180.0 180.0 (0.0) 180.0 (0.0)
C1 - C'2 - N4 - C6 0.0 0.0 (0.0) 0.0 (0.0)
O'3 - C'2 - N4 - C5 0.0 0.0 (0.0) 0.0 (0.0)
O'3 - C'2 - N4 - C6 180.0 180.0 (0.0) 180.0 (0.0)
C'2 - N4 - C5 - H10 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N4 - C5 - H11 120.3 120.7 (0.4) 119.8 (-0.5)
C'2 - N4 - C5 - H12 239.7 239.3 (-0.4) 240.2 (0.5)
C6 - N4 - C5 - H10 180.0 180.0 (0.0) 180.0 (0.0)
C6 - N4 - C5 - H11 300.3 300.7 (0.4) 299.8 (-0.5)
C6 - N4 - C5 - H12 59.7 59.3 (-0.4) 60.2 (0.5)
C'2 - N4 - C6 - H13 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N4 - C6 - H14 120.4 120.9 (0.5) 119.9 (-0.5)
C'2 - N4 - C6 - H15 239.6 239.1 (-0.5) 240.1 (0.5)
C5 - N4 - C6 - H13 180.0 180.0 (0.0) 180.0 (0.0)
C5 - N4 - C6 - H14 300.4 300.9 (0.5) 299.9 (-0.5)
C5 - N4 - C6 - H15 59.6 59.1 (-0.5) 60.1 (0.5)
Out-of-Planes (degrees)
C'2 - N4 - C5 - C6 0.0 0.0 (0.0) 0.0 (0.0)
Table II.G. Comparison of Urea Structures Optimized by Ab Initio Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
Bonds (Å)
N1 - C'2 1.373 1.308 (-0.065) 1.372 (-0.001)
N1 - H*5 0.996 0.996 (0.000) 0.996 (0.000)
N1 - H*6 0.996 0.994 (-0.003) 0.993 (-0.003)
C'2 - O'3 1.197 1.180 (-0.017) 1.192 (-0.005)
C'2 - N4 1.373 1.308 (-0.065) 1.372 (-0.001)
N4 - H*7 0.996 0.996 (0.000) 0.996 (0.000)
N4 - H*8 0.996 0.994 (-0.003) 0.993 (-0.003)
Angles (degrees)
C'2 - N1 - H*5 113.4 112.3 (-1.0) 109.3 (-4.1)
C'2 - N1 - H*6 117.7 114.5 (-3.3) 117.3 (-0.4)
H*5 - N1 - H*6 114.4 116.2 (1.7) 116.2 (1.8)
N1 - C'2 - O'3 123.0 122.5 (-0.5) 121.9 (-1.1)
N1 - C'2 - N4 114.0 115.0 (1.1) 116.2 (2.2)
O'3 - C'2 - N4 123.0 122.5 (-0.5) 121.9 (-1.1)
C'2 - N4 - H*7 113.4 112.3 (-1.0) 109.3 (-4.1)
C'2 - N4 - H*8 117.7 114.5 (-3.3) 117.3 (-0.4)
H*7 - N4 - H*8 114.4 116.2 (1.7) 116.2 (1.8)
Torsions (degrees)
H*5 - N1 - C'2 - O'3 12.6 13.9 (1.3) 9.2 (-3.4)
H*5 - N1 - C'2 - N4 192.6 193.9 (1.3) 189.2 (-3.4)
H*6 - N1 - C'2 - O'3 150.0 149.2 (-0.8) 144.2 (-5.8)
H*6 - N1 - C'2 - N4 330.0 329.2 (-0.8) 324.2 (-5.8)
N1 - C'2 - N4 - H*7 192.6 193.9 (1.3) 189.2 (-3.4)
N1 - C'2 - N4 - H*8 330.0 329.2 (-0.8) 324.2 (-5.8)
O'3 - C'2 - N4 - H*7 12.6 13.9 (1.3) 9.2 (-3.4)
O'3 - C'2 - N4 - H*8 150.0 149.2 (-0.8) 144.2 (-5.8)
Out-of Planes (degrees)
C'2 - N1 - H*6 - H*5 -37.4 -40.6 (-3.2) -40.8 (-3.3)
N1 - C'2 - O'3 - N4 0.0 0.0 (0.0) 0.0 (0.0)
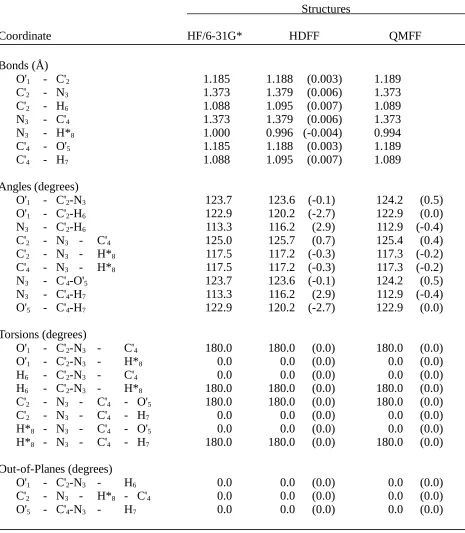
Table II.H. Comparison of N-Formylformamide Structures Optimizedby Ab Initio Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
Bonds (Å)
O'1 - C'2 1.185 1.188 (0.003) 1.189
C'2 - N3 1.373 1.379 (0.006) 1.373
C'2 - H6 1.088 1.095 (0.007) 1.089
N3 - C'4 1.373 1.379 (0.006) 1.373
N3 - H*8 1.000 0.996 (-0.004) 0.994
C'4 - O'5 1.185 1.188 (0.003) 1.189
C'4 - H7 1.088 1.095 (0.007) 1.089
Angles (degrees)
O'1 - C'2-N3 123.7 123.6 (-0.1) 124.2 (0.5)
O'1 - C'2-H6 122.9 120.2 (-2.7) 122.9 (0.0)
N3 - C'2-H6 113.3 116.2 (2.9) 112.9 (-0.4)
C'2 - N3 - C'4 125.0 125.7 (0.7) 125.4 (0.4)
C'2 - N3 - H*8 117.5 117.2 (-0.3) 117.3 (-0.2)
C'4 - N3 - H*8 117.5 117.2 (-0.3) 117.3 (-0.2)
N3 - C'4-O'5 123.7 123.6 (-0.1) 124.2 (0.5)
N3 - C'4-H7 113.3 116.2 (2.9) 112.9 (-0.4)
O'5 - C'4-H7 122.9 120.2 (-2.7) 122.9 (0.0)
Torsions (degrees)
O'1 - C'2-N3 - C'4 180.0 180.0 (0.0) 180.0 (0.0)
O'1 - C'2-N3 - H*8 0.0 0.0 (0.0) 0.0 (0.0)
H6 - C'2-N3 - C'4 0.0 0.0 (0.0) 0.0 (0.0)
H6 - C'2-N3 - H*8 180.0 180.0 (0.0) 180.0 (0.0)
C'2 - N3 - C'4 - O'5 180.0 180.0 (0.0) 180.0 (0.0)
C'2 - N3 - C'4 - H7 0.0 0.0 (0.0) 0.0 (0.0)
H*8 - N3 - C'4 - O'5 0.0 0.0 (0.0) 0.0 (0.0)
H*8 - N3 - C'4 - H7 180.0 180.0 (0.0) 180.0 (0.0)
Out-of-Planes (degrees)
O'1 - C'2-N3 - H6 0.0 0.0 (0.0) 0.0 (0.0)
C'2 - N3 - H*8 - C'4 0.0 0.0 (0.0) 0.0 (0.0)
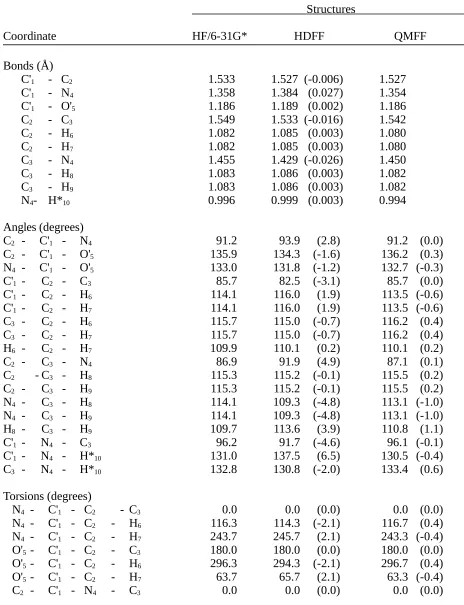
Table II.I. Comparison of Butyrolactam Structures Optimized by Ab Initio Calculation (HF/6-31G*) and by the HDFF and QMFF Force Fields.
Structures
Coordinate HF/6-31G* HDFF QMFF
Bonds (Å)
C'1 - C2 1.533 1.527 (-0.006) 1.527
C'1 - N4 1.358 1.384 (0.027) 1.354
C'1 - O'5 1.186 1.189 (0.002) 1.186
C2 - C3 1.549 1.533 (-0.016) 1.542
C2 - H6 1.082 1.085 (0.003) 1.080
C2 - H7 1.082 1.085 (0.003) 1.080
C3 - N4 1.455 1.429 (-0.026) 1.450
C3 - H8 1.083 1.086 (0.003) 1.082
C3 - H9 1.083 1.086 (0.003) 1.082
N4- H*10 0.996 0.999 (0.003) 0.994
Angles (degrees)
C2 - C'1 - N4 91.2 93.9 (2.8) 91.2 (0.0)
C2 - C'1 - O'5 135.9 134.3 (-1.6) 136.2 (0.3)
N4 - C'1 - O'5 133.0 131.8 (-1.2) 132.7 (-0.3)
C'1 - C2 - C3 85.7 82.5 (-3.1) 85.7 (0.0)
C'1 - C2 - H6 114.1 116.0 (1.9) 113.5 (-0.6)
C'1 - C2 - H7 114.1 116.0 (1.9) 113.5 (-0.6)
C3 - C2 - H6 115.7 115.0 (-0.7) 116.2 (0.4)
C3 - C2 - H7 115.7 115.0 (-0.7) 116.2 (0.4)
H6 - C2 - H7 109.9 110.1 (0.2) 110.1 (0.2)
C2 - C3 - N4 86.9 91.9 (4.9) 87.1 (0.1)
C2 - C3 - H8 115.3 115.2 (-0.1) 115.5 (0.2)
C2 - C3 - H9 115.3 115.2 (-0.1) 115.5 (0.2)
N4 - C3 - H8 114.1 109.3 (-4.8) 113.1 (-1.0)
N4 - C3 - H9 114.1 109.3 (-4.8) 113.1 (-1.0)
H8 - C3 - H9 109.7 113.6 (3.9) 110.8 (1.1)
C'1 - N4 - C3 96.2 91.7 (-4.6) 96.1 (-0.1)
C'1 - N4 - H*10 131.0 137.5 (6.5) 130.5 (-0.4)
C3 - N4 - H*10 132.8 130.8 (-2.0) 133.4 (0.6)
Torsions (degrees)
N4 - C'1 - C2 - C3 0.0 0.0 (0.0) 0.0 (0.0)
N4 - C'1 - C2 - H6 116.3 114.3 (-2.1) 116.7 (0.4)
N4 - C'1 - C2 - H7 243.7 245.7 (2.1) 243.3 (-0.4)
O'5 - C'1 - C2 - C3 180.0 180.0 (0.0) 180.0 (0.0)
O'5 - C'1 - C2 - H6 296.3 294.3 (-2.1) 296.7 (0.4)
O'5 - C'1 - C2 - H7 63.7 65.7 (2.1) 63.3 (-0.4)
C2 - C'1 - N4 - H*10 180.0 180.0 (0.0) 180.0 (0.0)
O'5 - C'1 - N4 - C3 180.0 180.0 (0.0) 180.0 (0.0)
O'5 - C'1 - N4 - H*10 0.0 0.0 (0.0) 0.0 (0.0)
C'1 - C2 - C3 - N4 0.0 0.0 (0.0) 0.0 (0.0)
C'1 - C2 - C3 - H8 244.7 247.6 (2.9) 245.8 (1.1)
C'1 - C2 - C3 - H9 115.3 112.4 (-2.9) 114.2 (-1.1)
H6 - C2 - C3 - N4 245.3 244.7 (-0.6) 245.9 (0.6)
H6 - C2 - C3 - H8 130.0 132.3 (2.3) 131.7 (1.7)
H6 - C2 - C3 - H9 0.6 357.1 (-3.5) 0.2 (-0.4)
H7 - C2 - C3 - N4 114.7 115.3 (0.6) 114.1 (-0.6)
H7 - C2 - C3 - H8 359.4 2.9 (3.5) 359.8 (0.4)
H7 - C2 - C3 - H9 230.0 227.7 (-2.3) 228.3 (-1.7)
C2 - C3 - N4 - C'1 0.0 0.0 (0.0) 0.0 (0.0)
C2 - C3 - N4 - H*10 180.0 180.0 (0.0) 180.0 (0.0)
H8 - C3 - N4 - C'1 116.4 117.6 (1.1) 116.5 (0.1)
H8 - C3 - N4 - H*10 296.4 297.6 (1.1) 296.5 (0.1)
H9 - C3 - N4 - C'1 243.6 242.4 (-1.1) 243.5 (-0.1)
H9 - C3 - N4 - H*10 63.6 62.4 (-1.1) 63.5 (-0.1)
Out-of-Planes (degrees)
C2 - C'1 - N4 - O'5 0.0 0.0 (0.0) 0.0 (0.0)
C'1 - N4 - C3 - H*10 0.0 0.0 (0.0) 0.0 (0.0)
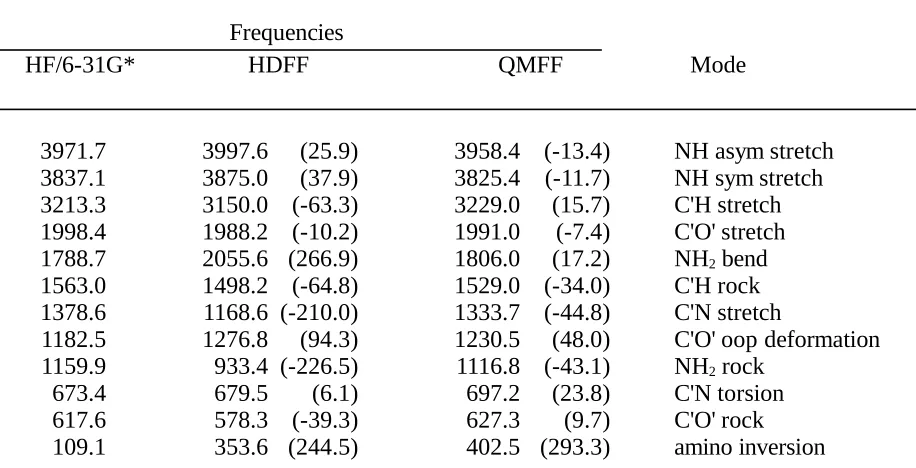
Table III.A. Comparison of Formamide Frequencies Obtained by Ab Initio Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3971.7 3997.6 (25.9) 3958.4 (-13.4) NH asym stretch
3837.1 3875.0 (37.9) 3825.4 (-11.7) NH sym stretch
3213.3 3150.0 (-63.3) 3229.0 (15.7) C'H stretch
1998.4 1988.2 (-10.2) 1991.0 (-7.4) C'O' stretch
1788.7 2055.6 (266.9) 1806.0 (17.2) NH2 bend
1563.0 1498.2 (-64.8) 1529.0 (-34.0) C'H rock
1378.6 1168.6 (-210.0) 1333.7 (-44.8) C'N stretch
1182.5 1276.8 (94.3) 1230.5 (48.0) C'O' oop deformation
1159.9 933.4 (-226.5) 1116.8 (-43.1) NH2 rock
673.4 679.5 (6.1) 697.2 (23.8) C'N torsion
617.6 578.3 (-39.3) 627.3 (9.7) C'O' rock
Table III.B. Comparison of Planar Acetamide Frequencies Obtained by Ab Initio Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3975.1 3997.6 (22.5) 3970.0 (-5.1) NH asym stretch
3843.4 3873.9 (30.6) 3829.1 (-14.3) NH sym stretch
3337.2 3268.7 (-68.5) 3308.6 (-28.7) CH3 asym stretch
3272.1 3259.2 (-12.9) 3265.5 (-6.5) CH3 asym stretch
3214.4 3140.7 (-73.7) 3226.7 (12.3) CH3 sym stretch
1985.2 1974.8 (-10.4) 2004.3 (19.0) C'O' stretch
1795.0 2049.8 (254.8) 1799.8 (4.9) NH2 bend
1627.2 1624.8 (-2.4) 1692.9 (65.7) CH3 asym deformation
1617.2 1623.3 (6.1) 1681.7 (64.5) CH3 asym deformation
1562.3 1543.3 (-19.0) 1619.7 (57.5) CH3 sym deformation
1463.6 1324.0 (-139.6) 1485.5 (21.9) C'N stretch
1234.2 1083.8 (-150.4) 1151.6 (-82.7) NH2 rock
1168.9 1042.5 (-126.4) 1143.8 (-25.1) CH3 rock
1076.6 1001.5 (-75.1) 1060.5 (-16.1) CH3 rock
913.0 733.5 (-179.5) 886.4 (-26.6) C'C stretch
696.4 783.9 (87.5) 715.0 (18.5) C'O' oop deformation
598.1 558.2 (-39.9) 605.7 (7.6) C'O' rock
562.6 641.6 (79.0) 614.1 (51.5) C'N torsion
446.6 356.5 (-90.1) 403.7 (-42.9) C'C rock
11.7 -6.5a,b (-18.2) 118.0 (106.3) CH
3 torsion
-144.5a 175.9 (320.4) 380.8 (525.3) amino inversion
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue. bThis imaginary frequency arises from selecting the force field stationary state with the geometry closest to the corresponding ab initio structure for comparison. In this case the configuration computed with the force field corresponds to a transition state.
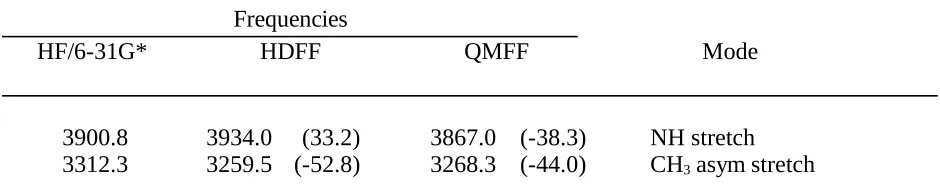
Table III.C. Comparison of Planar Trans N-Methylformamide Frequencies Obtained by Ab Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3900.8 3934.0 (33.2) 3867.0 (-38.3) NH stretch
3296.9 3262.8 (-34.1) 3255.2 (-41.7) CH3 asym stretch
3230.5 3142.0 (-88.6) 3207.0 (-23.5) CH3 sym stretch
3206.7 3149.2 (-57.5) 3235.7 (28.9) C'H stretch
1977.2 1994.6 (17.4) 1978.5 (1.3) C'O' Stretch
1694.9 1792.2 (97.3) 1699.2 (4.2) NH rock
1655.1 1617.8 (-37.3) 1653.2 (-1.9) CH3 asym deformation
1629.7 1605.9 (-23.8) 1637.8 (8.1) CH3 asym deformation
1609.3 1586.9 (-22.4) 1601.2 (-8.2) CH3 sym deformation
1559.9 1487.6 (-72.3) 1529.4 (-30.5) C'H rock
1348.6 1113.1 (-235.4) 1304.3 (-44.2) C'N stretch
1283.7 1039.3 (-244.4) 1239.1 (-44.6) CH3 rock
1266.0 1034.3 (-231.7) 1269.5 (3.5) CH3 rock
1170.7 1283.5 (112.8) 1242.8 (72.1) C'O' oop deformation
1035.4 823.9 (-211.5) 1000.1 (-35.3) NC stretch
837.8 692.0 (-145.9) 812.8 (-25.0) C'O' rock
486.9 560.8 (73.9) 574.4 (87.5) C'N torsion
291.8 301.2 (9.4) 317.4 (25.6) NC rock
228.5 323.4 (94.9) 328.1 (99.6) amino inversion
-60.6a -62.5a (-1.9) -136.5a (-75.9) CH
3 torsion
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue.
Table III.D. Comparison of Cis N-Methylformamide Frequencies Obtained by Ab Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3855.6 3941.0 (85.4) 3810.5 (-45.1) NH stretch
3299.0 3259.9 (-39.0) 3278.2 (-20.8) CH3 asym stretch
3268.8 3261.4 (-7.4) 3252.0 (-16.8) CH3 asym stretch
3215.4 3141.7 (-73.6) 3206.4 (-8.9) CH3 sym stretch
3201.4 3149.8 (-51.6) 3205.7 (4.3) C'H stretch
1988.8 2012.1 (23.3) 1988.0 (-0.8) C'O' stretch
1681.4 1616.1 (-65.3) 1690.8 (9.4) CH3 asym deformation
1643.2 1748.2 (105.0) 1626.3 (-16.9) NH rock
1631.5 1602.2 (-29.3) 1641.2 (9.7) CH3 asym deformation
1623.1 1595.2 (-27.9) 1638.1 (15.0) CH3 sym deformation
1542.4 1515.9 (-26.5) 1562.4 (20.1) C'H rock
1412.8 1049.9 (-362.9) 1386.1 (-26.7) C'N stretch
1270.2 1105.5 (-164.7) 1241.8 (-28.3) CH3 rock
1258.9 1024.2 (-234.7) 1241.0 (-18.0) CH3 rock
1179.5 1254.5 (75.1) 1180.0 (0.5) C'O' oop deformation
1096.1 953.9 (-142.1) 1068.9 (-27.1) NC stretch
648.6 631.0 (-17.6) 698.3 (49.7) C'N torsion
374.6 343.6 (-31.0) 345.5 (-29.1) NC rock
193.0 179.5 (-13.5) 215.3 (22.3) CH3 torsion
66.3 -50.8a (-117.1) -74.9a (-141.2) amino inversion
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue.
Table III.E. Comparison of Trans N-Methylacetamide Frequencies Obtained by Ab Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3914.8 3934.0 (19.2) 3886.2 (-28.6) NH stretch
3307.6 3259.5 (-48.1) 3269.1 (-38.4) N-CH3 asym stretch
3301.9 3261.6 (-40.3) 3267.1 (-34.9) C'-CH3 asym stretch
3296.4 3265.0 (-31.4) 3280.5 (-15.9) C'-CH3 asym stretch
3295.9 3262.9 (-33.0) 3256.2 (-39.7) N-CH3 asym stretch
3229.0 3142.0 (-87.0) 3207.7 (-21.3) N-CH3 sym stretch
3223.7 3140.9 (-82.8) 3222.0 (-1.6) C'-CH3 sym stretch
1956.5 1974.9 (18.4) 1973.3 (16.7) C'O' stretch
1711.0 1792.0 (81.0) 1707.0 (-4.0) NH rock
1658.9 1617.7 (-41.2) 1675.2 (16.2) N-CH3 asym deformation
1630.8 1627.0 (-3.8) 1650.4 (19.6) C'-CH3 asym deformation
1628.8 1605.9 (-22.9) 1639.8 (11.0) N-CH3 asym deformation
1617.3 1626.3 (9.0) 1693.2 (75.9) C'-CH3 asym deformation
1609.1 1586.3 (-22.8) 1603.4 (-5.7) N-CH3 sym deformation
1555.2 1542.0 (-13.2) 1594.6 (39.4) C'-CH3 sym deformation
1408.2 1289.9 (-118.3) 1416.8 (8.6) C'-N stretch
1311.3 1044.3 (-267.0) 1249.8 (-61.4) N-CH3 rock
1265.7 1030.5 (-235.2) 1268.7 (3.0) N-CH3 rock
1191.5 1007.8 (-183.7) 1116.5 (-75.0) N-C stretch
1172.2 1058.3 (-113.9) 1151.6 (-20.6) C'-CH3 rock
1094.9 1034.8 (-60.1) 1033.7 (-61.2) C'-CH3 rock
950.6 731.2 (-219.4) 932.8 (-17.8) C'-C stretch
686.0 793.6 (107.6) 714.4 (28.4) C'O' oop deformation
673.6 605.8 (-67.8) 653.6 (-20.0) C'O' rock
456.5 381.9 (-74.6) 426.7 (-29.9) C'C rock
365.9 373.4 (7.5) 506.5 (140.6) C'N torsion
284.0 266.6 (-17.4) 290.8 (6.8) NC rock
168.1 206.6 (38.5) 190.2 (22.1) amino inversion
19.8 -55.5a,b (-75.3) -110.4a,b(-130.2) C'-CH
3 torsion -41.4a -78.5a (-37.1) -127.0a (-85.6) N-CH
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue. bThis imaginary frequency arises from selecting the force field stationary state with the geometry closest to the corresponding ab initio structure for comparison. In this case the configuration computed with the force field corresponds to a transition state.
Table III.F. Comparison of Cis Planar N-Methylacetamide Frequencies Obtained byAb Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3872.7 3940.6 (67.9) 3831.3 (-41.4) NH stretch
3337.8 3268.6 (-69.2) 3305.8 (-32.0) C'-CH3 asym stretch
3306.7 3258.8 (-47.9) 3263.7 (-43.1) N-CH3 asym stretch
3276.5 3258.6 (-17.9) 3267.2 (-9.3) C'-CH3 asym stretch
3260.8 3262.6 (1.8) 3259.1 (-1.8) N-CH3 asym stretch
3218.9 3140.4 (-78.5) 3227.3 (8.4) C'-CH3 sym stretch
3208.6 3141.8 (-66.8) 3208.0 (-0.6) N-CH3 sym stretch
1969.9 1985.1 (15.2) 1999.5 (29.6) C'O' stretch
1670.8 1620.2 (-50.6) 1694.5 (23.7) N-CH3 asym deformation
1644.2 1608.4 (-35.8) 1643.0 (-1.2) N-CH3 asym deformation
1633.9 1583.5 (-50.4) 1634.7 (0.8) N-CH3 sym deformation
1626.7 1742.4 (115.7) 1589.0 (-37.7) NH rock
1625.2 1627.7 (2.5) 1701.9 (76.7) C'-CH3 asym deformation
1611.2 1628.6 (17.4) 1686.0 (74.8) C'-CH3 asym deformation
1562.4 1543.6 (-18.8) 1617.5 (55.1) C'-CH3 deformation
1469.5 1314.4 (-155.1) 1492.6 (23.1) C'-N stretch
1318.6 1056.7 (-261.9) 1242.7 (-75.9) N-CH3 rock
1260.9 1031.9 (-229.0) 1264.9 (4.0) N-CH3 rock
1193.1 1029.5 (-163.6) 1131.4 (-61.7) NC stretch
1166.8 1042.9 (-123.9) 1137.5 (-29.3) C'-CH3 rock
1093.9 1003.4 (-90.5) 1077.4 (-16.4) C'-CH3 rock
867.3 708.5 (-158.8) 838.7 (-28.6) C'C stretch
671.0 771.7 (100.7) 688.5 (17.4) C'O' oop deformation
611.9 457.7 (-154.2) 598.0 (-14.0) C'O' rock
529.0 507.0 (-22.0) 495.1 (-33.9) C'C rock
508.3 594.3 (86.0) 477.9 (-30.4) C'N torsion
297.8 268.3 (-29.5) 295.0 (-2.7) NC rock
183.4 142.1 (-41.3) 195.1 (11.7) N-CH3 torsion
115.2 98.5 (-16.7) 166.1 (50.9) C'-CH3 torsion
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue.
Table III.G. Comparison of N,N-Dimethylformamide Frequencies Obtained by Ab Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3361.4 3261.1 (-100.3) 3293.8 (-67.7) CH3 asym stretch
3300.4 3264.4 (-36.0) 3290.6 (-9.8) CH3 asym stretch
3249.6 3259.4 (9.8) 3233.1 (-16.5) CH3 asym stretch
3248.6 3259.0 (10.4) 3234.2 (-14.4) CH3 asym stretch
3214.5 3148.5 (-66.0) 3208.6 (-5.8) C'H stretch
3201.1 3142.7 (-58.4) 3194.5 (-6.6) CH3 sym stretch
3196.2 3143.5 (-52.7) 3193.1 (-3.1) CH3 sym stretch
1961.1 2001.1 (40.0) 1974.2 (13.1) C'O' stretch
1686.6 1594.5 (-92.1) 1698.2 (11.5) CH3 asym deformation
1655.8 1609.2 (-46.6) 1689.6 (33.8) CH3 asym deformation
1647.7 1621.2 (-26.5) 1648.1 (0.4) CH3 asym deformation
1627.0 1614.2 (-12.8) 1642.5 (15.5) CH3 asym deformation
1626.7 1641.3 (14.6) 1666.4 (39.7) CH3 sym deformation
1592.2 1654.0 (61.8) 1670.9 (78.7) CH3 sym deformation
1586.8 1284.7 (-302.1) 1601.8 (14.9) C'N stretch
1557.1 1525.0 (-32.1) 1553.1 (-4.0) C'H rock
1404.6 1231.3 (-173.3) 1406.0 (1.4) NC asym stretch
1287.8 1042.6 (-245.2) 1273.9 (-13.9) CH3 rock
1239.0 1001.6 (-237.4) 1225.2 (-13.8) CH3 rock
1199.3 988.7 (-210.6) 1160.2 (-39.2) CH3 rock
1185.5 1028.8 (-156.7) 1180.7 (-4.8) CH3 rock
1165.1 1262.6 (97.5) 1187.1 (22.0) C'O' oop deformation
941.5 821.0 (-120.5) 909.7 (-31.8) NC sym stretch
711.9 586.9 (-125.0) 695.7 (- 16.2) C'O' rock
426.3 419.6 (-6.7) 402.1 (-24.2) CNC bend
351.2 307.3 (-43.9) 356.3 (5.1) CNC rock
346.1 394.9 (48.8) 352.0 (6.0) C'N torsion
242.7 258.8 (16.1) 243.1 (0.4) amino inversion
161.7 143.3 (-18.4) 83.4 (-78.2) CH3 sym torsion
83.9 87.8 (3.9) -127.2a,b (-211.1) CH
3 asym torsion
the geometry closest to the corresponding ab initio structure for comparison. In this case the configuration computed with the force field corresponds to a transition state.
Table III.H. Comparison of Planar N,N-Dimethylacetamide Frequencies Obtained by Ab Initio
Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3379.0 3261.1 (-118.0) 3300.1 (-79.0) N-CH3 asym stretch
3357.1 3265.0 (-92.1) 3321.4 (-35.7) N-CH3 asym stretch
3338.9 3267.8 (-71.1) 3299.4 (-39.4) C'-CH3 asym stretch
3281.7 3258.1 (-23.6) 3267.4 (-14.3) C'-CH3 asym stretch
3241.3 3259.1 (17.8) 3228.7 (-12.6) N-CH3 asym stretch
3240.3 3258.2 (17.9) 3233.0 (-7.3) N-CH3 asym stretch
3221.5 3140.1 (-81.4) 3226.5 (5.0) C'-CH3 sym stretch
3203.2 3143.8 (-59.4) 3194.4 (-8.8) C'-CH3 sym stretch
3193.5 3145.3 (-48.2) 3191.9 (-1.6) N-CH3 sym stretch
1935.2 1968.4 (33.3) 1974.6 (39.4) C'O' stretch
1690.9 1592.8 (-98.1) 1675.7 (-15.3) N-CH3 asym deformation
1657.5 1605.0 (-52.5) 1679.5 (22.0) N-CH3 asym deformation
1655.2 1619.7 (-35.5) 1647.8 (-7.4) N-CH3 asym deformation
1645.4 1633.5 (-11.9) 1640.0 (-5.5) N-CH3 asym deformation
1637.8 1669.2 (31.4) 1710.1 (72.3) N-CH3 sym deformation
1627.7 1610.9 (-16.8) 1703.5 (75.9) C'-CH3 asym deformation
1616.3 1633.6 (17.3) 1703.3 (87.0) C'-CH3 asym deformation
1593.0 1371.7 (-221.3) 1621.9 (28.9) N-CH3 sym deformation
1581.6 1675.5 (93.9) 1690.5 (108.9) C'-N stretch
1538.5 1549.1 (10.6) 1569.1 (30.6) C'-CH3 sym deformation
1408.1 1242.1 (-166.0) 1423.2 (15.1) NC asym stretch
1313.6 1156.7 (-156.9) 1302.0 (-11.6) N-CH3 rock
1286.7 1034.1 (-252.6) 1269.5 (-17.1) N-CH3 rock
1237.5 997.6 (-239.8) 1219.5 (-18.0) N-CH3 rock
1182.2 1040.0 (-142.2) 1184.7 (2.5) N-CH3 rock
1164.0 1053.9 (-110.1) 1129.3 (-34.7) C'-CH3 rock
1126.8 1028.2 (-98.6) 1109.2 (-17.7) C'-CH3 rock
1053.6 966.5 (-87.1) 1021.5 (-32.2) NC sym stretch
789.1 678.5 (-110.7) 767.4 (-21.7) C'C stretch
651.2 782.8 (131.6) 679.2 (28.0) C'O' oop deformation
639.1 564.8 (-74.2) 635.7 (-3.4) C'O' rock
500.7 390.0 (-110.8) 470.9 (-29.8) C'C rock
435.6 439.9 (4.3) 435.7 (0.2) CNC bend
348.3 321.6 (-26.7) 361.0 (12.8) CNC rock
212.5 208.1 (-4.4) 230.0 (17.5) C'-CH3 torsion
143.1 145.8 (2.7) 128.7 (-14.5) C'N torsion
78.3 85.0 (6.7) -84.1a,b (-162.5) N-CH
3 torsion -62.8a 10.9 (73.7) -171.2a (-108.4) N-CH
3 torsion
a The minus sign indicates that the frequency was calculated from the absolute value of a negative eigenvalue. bThis imaginary frequency arises from selecting the force field stationary state with the geometry closest to the corresponding ab initio structure for comparison. In this case the configuration computed with the force field corresponds to a transition state.
Table III.I. Comparison of Urea Frequencies Obtained by Ab Initio Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3926.2 3989.0 (62.8) 3927.2 (1.0) NH asym stretch
3925.9 3996.5 (70.6) 3932.8 (6.9) NH asym stretch
3814.9 3884.8 (69.9) 3813.5 (-1.4) NH sym stretch
3810.8 3870.7 (59.9) 3801.9 (-8.9) NH sym stretch
2000.3 2037.0 (36.7) 1996.5 (-3.7) C'O' stretch
1811.9 1929.0 (117.1) 1726.5 (-85.4) NH2 asym bend
1805.5 1953.0 (147.5) 1755.0 (-50.5) NH2 sym bend
1550.2 1404.7 (-145.5) 1552.1 (1.8) C'N asym stretch
1303.6 1118.7 (-184.9) 1168.4 (-135.2) NH2 sym rock
1166.1 990.0 (-176.1) 1064.0 (-102.1) NH2 asym rock
1034.8 640.8 (-394.0) 947.2 (-87.6) C'N sym stretch
882.1 919.3 (37.2) 970.8 (88.7) C'O' oop deformation
648.8 770.6 (121.8) 720.7 (71.9) asym amino inversion
629.9 541.3 (-88.6) 666.3 (36.4) C'O' rock
598.8 675.5 (76.7) 607.6 (8.8) sym amino inversion
514.2 498.3 (-15.9) 567.5 (53.3) NC'N bend
483.2 415.1 (-68.1) 435.5 (-47.6) asym C'N torsion
417.2 416.5 (-0.7) 452.3 (35.1) sym C'N torsion
Table III.J. Comparison of N-Formylformamide Frequencies Obtained by Ab Initio Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3256.1 3152.0 (-104.1) 3228.4 (-27.7) C'H sym stretch
3232.7 3148.7 (-84.0) 3224.4 (-8.2) C'H asym stretch
2046.2 1993.8 (-52.4) 2010.4 (-35.8) C'O' sym stretch
2007.6 2017.5 (9.9) 1996.8 (-10.8) C'O asym stretch
1627.9 1534.7 (-93.2) 1570.2 (-57.7) NH rock
1584.9 1518.6 (-66.3) 1575.8 (-9.1) C'H sym rock
1488.2 1493.6 (5.4) 1483.4 (-4.9) C'H asym rock
1370.2 1133.2 (-237.0) 1311.5 (-58.7) C'N sym stretch
1294.4 992.4 (-302.0) 1305.6 (11.2) C'N asym stretch
1176.5 1240.9 (64.4) 1197.2 (20.7) C'O' asym oop deformation
1161.9 1220.6 (58.7) 1163.6 (1.7) C'O' sym oop deformation
777.1 856.5 (79.4) 803.6 (26.5) C'N asym torsion
687.2 680.5 (-6.7) 721.7 (34.5) C'O' asym rock
580.1 430.3 (-149.8) 570.7 (-9.4) C'O' sym rock
303.1 306.7 (3.6) 299.2 (-3.9) C'N sym torsion
275.7 301.4 (25.7) 325.9 (50.2) C'NC' bend
137.3 145.6 (8.3) 142.7 (5.4) amino inversion
Table III.K. Comparison of Butyrolactam Frequencies Obtained by Ab Initio Calculation (HF/6-31G*) and the HDFF and QMFF Force Fields.
Frequencies
HF/6-31G* HDFF QMFF Mode
3878.3 3937.5 (59.2) 3893.3 (15.0) NH stretch
3324.1 3267.3 (-56.8) 3318.1 (-5.9) CH asym stretch
3282.5 3276.7 (-5.8) 3295.6 (13.1) CH asym stretch
3268.7 3187.2 (-81.5) 3278.1 (9.4) CH sym stretch
3238.4 3175.4 (-62.9) 3247.6 (9.3) CH sym stretch
2057.2 2013.5 (-43.7) 2072.5 (15.3) C'O' stretch
1684.3 1705.0 (20.7) 1709.9 (25.6) CH2 scissor
1609.6 1647.6 (37.9) 1685.9 (76.2) CH2 scissor
1548.7 1203.2 (-345.5) 1449.2 (-99.5) NH rock
1457.9 1345.6 (-112.2) 1431.7 (-26.2) CH2 wag
1353.7 1136.7 (-217.0) 1186.9 (-166.8) CH2 wag
1310.9 1027.4 (-283.5) 1357.4 (46.5) CH2 twist
1304.7 955.5 (-349.1) 1291.6 (-13.0) C'N stretch
1264.2 1088.0 (-176.2) 1270.6 (6.4) CH2 twist
1170.5 1142.2 (-28.3) 1121.1 (-49.4) CH2 rock
1099.4 1783.6 (684.3) 1057.9 (-41.4) NC stretch
1056.5 862.4 (-194.1) 1095.9 (39.4) C'C stretch
965.5 774.6 (-190.9) 913.0 (-52.5) CC stretch
896.7 891.3 (-5.4) 906.5 (9.8) CH2 rock
817.1 519.6 (-297.4) 674.4 (-142.6) ring deformation
518.5 554.4 (35.9) 530.4 (11.9) C'O' rock
407.6 -498.6a (-906.2) 556.5 (148.9) ring puckering
75.3 366.1 (290.7) 199.7 (124.4) amino inversion