Cloning and characterization of a cDNA encoding hexokinase
from tomato
Thierry Menu
a, Christophe Rothan
a, Nir Dai
b, Marina Petreikov
b,
Christelle Etienne
a, Agnes Destrac-Irvine
a, Arthur Schaffer
b, David Granot
b,
Be´re´nice Ricard
a,*
aUnite´ de Physiologie Ve´ge´tale,Institut National de la Recherche Agronomique,B.P.81,33883Villena6e d’Ornon,France bInstitute of Field and Garden Crops,Agricultural Research Organization,Volcani Center,Bet Dagan50250,Israel
Received 17 April 2000; received in revised form 30 June 2000; accepted 30 June 2000
Abstract
Two different partial sequences encoding putative hexokinase (HXK, ATP: hexose-6-phosphotransferase, EC 2.7.1.1) were isolated from tomato (Lycopersicon esculentum) by RT-PCR using degenerate primers. Southern blot analysis suggested the existence of two divergent HXK genes. A complete cDNA of one HXK was isolated by screening a cDNA library prepared from young cherry tomato fruit. The 1770 bp cDNA ofLeHXK2 contained an open reading frame encoding a 496 amino acid protein that has 69% identity with the two Arabidopsis HXKs, 83 and 85% identity with potato StHXK1 and tobacco NtHXK, respectively. However, this clone had 97% amino acid identity with potatoStHXK2 and, therefore, was namedLeHXK2.LeHXK2 cDNA was expressed in a triple mutant yeast (Saccharomyces cere6isiae) strain which lacked the ability to phosphorylate glucose
and fructose and, therefore, was unable to grow on these sugars as carbon sources. Mutant cells expressingLeHXK2 grew on both glucose and fructose with shorter doubling time on glucose. The kinetic properties ofLeHXK2 expressed in yeast were determined after the purification ofLeHXK2 by HPLC-ion exchange chromatography, confirming the identity ofLeHXK2 as hexokinase with higher affinity to glucose.LeHXK2 mRNA was detected by RT-PCR expression analysis in all organs and tissues and at all stages of fruit development. However, semi-quantitative RT-PCR analysis showed thatLeHXK2 was most highly expressed in flowers. © 2001 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Sugar phosphorylation;Lycopersicon esculentum; cDNA
www.elsevier.com/locate/plantsci
1. Introduction
Hexose phosphorylating enzymes are function-ally classified as fructokinase (FRK), glucokinase (GLK) or hexokinase (HXK), based on their sub-strate specificities. FRK and GLK phosphorylate
strictly fructose (Fru) and glucose (Glc), respec-tively, while HXK can phosphorylate a range of hexoses (e.g. Fru, Glc and Man). Together, these enzymes catalyze the first, irreversible step of hexose metabolism. These reactions potentially constitute an important regulatory step in plants and in other organisms [1 – 3].
Only a few genes encoding hexose phosphory-lating enzymes have been cloned from plants in-cluding tomato. Two tomato cDNAs encoding FRK were isolated and their ability to comple-ment a yeast triple mutant unable to phosphory-late either Glc or Fru showed that both cDNAs encoded genuine FRK [4,5]. Several HXK genes
have been cloned so far, two from Arabidopsis
Abbre6iations:DAA, days after anthesis; FRK, fructokinase; Fru, fructose; Gal, galactose; Glc, Glucose; Glc 6-P, glucose 6-phosphate; GLK, glucokinase; HXK, hexokinase; Man, mannose; PGI, phos-phoglucose isomerase; PGK, phosphoglycerate kinase; RT, reverse transcrip(tase/tion); Suc, sucrose; Ura, uracil.
The nucleotide sequence ofLeHXK2will appear in the GenBank Nucleotide Sequence Database under the accession number AF208543.
* Corresponding author. Tel.: +33-55-6843234; fax: + 33-55-6843245.
E-mail address:[email protected] (B. Ricard).
(AtHXK1 andAtHXK2) and potato (StHXK1 and
StHXK2), one from tobacco and one from spinach ([1,6,7] and accession numbers X94302;
AF118133). TheArabidopsis HXKgenes were
pro-posed to be sugar sensors based on results
ob-tained with transgenic Arabidopsis plants
overexpressing AtHXK [1]. Similar results were
obtained with transgenic tomato plants
overex-pressing AtHXK suggesting that the sugar
signal-ing pathways in tomato might also be mediated by hexokinase [8]. To analyze the role of tomato hexokinase in tomato plants it was important to isolate and characterize tomato HXKs. In this paper, we describe the isolation of two different partial sequences of tomato HXKs that were used to demonstrate the existence of two divergent HXK genes in tomato plants. A full-length cDNA encoding one of the two HXKs was cloned and
designatedLeHXK2 based on its higher homology
to StHXK2. Kinetic analyses of LeHXK2 follow-ing expression in yeast cells suggest that LeHXK2 is not the GLK previously reported by Martinez-Barajas and Randall [9]. Expression analysis of
LeHXK2 indicates that LeHXK2 is expressed in a large spectrum of tissues and organs and at differ-ent stages of fruit developmdiffer-ent with the lowest expression in leaves and the highest in flowers.
2. Materials and methods
2.1. PCR amplification of cDNA and sequencing
Alignment of the predicted amino acid
se-quences of three plant HXK genes (Arabidopsis:
U28214, U28215 and Solanum tuberosum X94302)
identified several conserved domains. The domains corresponding to amino acids MTVEMHA and EMVINMEW were used to design the forward
(5%- ATG ACI GTI GAR ATG CAY GC -3%) and
the reverse (5%- CCA YTC CAT RTT DAT NAC
CAT YT -3%) degenerate primers, where I=
inosine, N=all the four nucleotides, D=A/G/T,
R=A/G, and Y=C/T. PCR was carried out with
cDNA from fruit 10 DAA. Amplifications were for 30 cycles, each consisting of 1 min at 95°C, 30 s at 50°C and 1 min 30 s at 72°C. The resulting approximately 700 bp fragment was purified on a S-400 MicrospinTM Column (Pharmacia Biotech)
and cloned into the pGEM®-T plasmid (Promega).
Four PCR products were partially sequenced
us-ing the Thermo Sequenase Cycle Sequencus-ing Kit (Amersham Life Science), revealing two different sequences that encoded putative HXK proteins.
2.2. cDNA isolation and characterization
An oligo dT-primed tomato fruit cDNA library
constructed in l Uni-ZAP XR [10] was separately
screened with the two partial HXK sequences. All the positive clones obtained corresponded to only one of the two partial sequences. The longest insert was commercially sequenced (MWG-Bio-tech, Germany) and the deduced amino acid se-quence aligned with other HXK sese-quences using [email protected].
2.3. Plasmids and yeast transformation
A yeast shuttle vector, pFL61, containing the
URA3 gene as a selective marker and the
constitu-tive phosphoglycerate kinase (PGK) promoter and terminator [11] was used to express the tomato
HXK cDNA clone in yeast cells. The cDNA was
inserted as a NotI/EcoRI fragment between the
NotI/EcoRI sites of pFL61; after verification, the
new plasmid was named pFL61-LeHXK2. The
yeast (Saccharomyces cere6isiae) strain used was
DFY632 — MATa, ura 3-52, hxk1::LEU2,
hxk2::LEU2, glk1::LEU2, lys 1-1, leu 2-1 [12].
Yeast transformations were carried out by grow-ing yeast cells in YEPG liquid medium, consistgrow-ing of 1% yeast extract (Difco), 2% Bacto Peptone (Difco) and 2% Gal, to mid-logarithmic phase, treating the cells with lithium acetate according to Ito et al. [13] and selecting for transformants on
−Ura+Gal plates. Selective medium for uracil
auxotrophic growth (−Ura+sugar) contained
0.5% ammonium sulfate, 0.17% yeast nitrogen base without amino acids (Difco), 0.2% casamino acids (Difco or Sigma), 0.004% adenine (Sigma), 0.008% Tryptophane (Sigma) and 2% Gal, Fru or Glc.
2.4. Protein purification of yeast expressed LeHXK2 and kinetic characterization
DFY632 yeast cells transformed with either
pFL61 or pFL-LeHXK2 were grown in minimal
by Kanayama et al. [4]. For the kinetic character-ization the 80% ammonium sulfate precipitate was collected, resuspended in extraction buffer and desalted on Sephadex G-25. The protein was applied to MonoQ (Pharmacia, 5 ml) preequili-brated with the extraction buffer but containing only 20 mM HEPES pH 7.0. Unbound protein was eluted with the same buffer, followed by a 0 – 0.5 M KCl salt gradient. Fractions of 0.5 ml were collected and Glc phosphorylating activities were measured as described below. One major peak of Glc phosphorylating activity was ob-served and the most active fractions (1 ml) were used as the partially purified enzyme for charac-terization.
HXK activity was measured according to Schaffer and Petreikov [14] for kinetic measure-ments or Bouny and Saglio [15] for inhibitor studies. Assays for kinetic measurements con-tained, in a total volume of 1 ml, 30 mM Hepes –
NaOH (pH 7.5), 1 mM MgCl2, 0.6 mM EDTA, 9
mM KCl, 1 mM NAD, 1 mM ATP, 2 U NAD-dependent Glc 6-P-dehydrogenase (G6PDH, Leu-conostoc). For the assay of Glc phosphorylation, the reaction was initiated with 2 mM Glc; for the assay of Fru phosphorylation, 2 U of PGI were added and the reaction was initiated with Fru.
Reactions were carried out at 37°C and the A340
was monitored continuously. Inhibitor studies were carried out in a similar fashion but con-tained 50 mM glucosamine or mannoheptulose or 5 mM ADP in 100 mM Tricine (pH 7.5), 2 mM
MgCl2, 3 mM DTT, 2 mM NAD, 2 mM ATP, 4
U G6PDH (Leuconostoc). Assays for Glc 6-P inhibition contained 5 mM Glc 6-P in 100 mM
Tricine (pH 7.5), 2 mM MgCl2, 3 mM DTT, 1.5
mM ATP, 150 mM NADH, 1.5 mM PEP, 3.6 U
PK, 16.6 U LDH. Reactions were initiated with 1 mM Glc and were carried out at 25°C.
2.5. DNA extraction and Southern blot analysis
Genomic DNA was extracted from green leaves of tomato plants according to Dellaporta
et al. [16]. DNA (10 mg) were totally digested
with different restriction enzymes, separated on 1.2% agarose gels, then transferred to nylon
membranes in 20×SSC according to standard
methods. Radiolabeling of cDNA probes was carried out by the random priming method using the Prime-It kit (Stratagene). Prehybridization
and hybridization reactions were carried
out at 65°C overnight in 6.6×SSC, 0.1% SDS,
5×Denhardt’s solution, 0.1% SDS at 65°C for
30 min.
2.6. RT-PCR expression analysis
Tissues were collected from plants grown under
controlled conditions (12 h day at 25°C/12 h
night at 20°C), frozen in liquid N2 and stored at
−80°C. Before extraction, tissues were reduced
to a powder under liquid N2. Total RNA was
then isolated by the hot phenol method described
by Verwoerd et al. [17]. Total RNA (2 mg) was
reverse transcribed and amplified using the Ac-cess RT-PCR System (Promega, France) and the
appropriate primers. To amplify LeHXK2, the
primers were S2 (5%- T AGC TAT GTA GAC
AAT CTC CCT- 3%) corresponding to amino
acids SYVDNL and AS1 (Fig. 2). To amplify Actin (Tom52 gene product, Genebank Accession
U60482) the primers were ActinS (5%- TGG CAT
CAT ACC TTT TAC AAT GAA- 3%) and
Acti-nAS (5%- CCT GAT ATC AAC GTC ACA CTT
CAT- 3%), corresponding to amino acids
WHHT-FYNE and MKCDVDIR. The positions of HXK primers are shown on the deduced amino acid sequences of Fig. 2. First strand cDNA reac-tions in which RT was inactivated by heating at 94°C for 2 min did not yield amplification prod-ucts.
The relative amounts of LeHXK2 mRNA were
determined via semi-quantitative RT-PCR in two independent experiments. The first strand cDNA
synthesis reactions were performed on 2 mg total
RNA which had been treated with RQ1 RNase-Free DNase (Promega) using an oligo-dT primer and MMLV RT as described in Moing et al. [18]. cDNA corresponding to 200 ng of total RNA was then PCR-amplified using the sense and anti-sense primers described above. Reactions were run on a Gene AmpTM PCR System 9600 (Perkin Elmers Applied Biosystems, France) for 30 cycles of 94°C (30 s), 50°C (30 s), 68°C (1 min) for HXK primers and 20 cycles for actin primers. These conditions were chosen so that amplification occurred in the linear range. PCR products were separated on 1% agarose gels and blotted onto nylon membranes which were then hybridized under stringent conditions with the
3. Results
3.1. Cloning of the full-length LeHXK2 cDNA
Total RNA extracted from tomato fruit har-vested 10 DAA was reverse transcribed. The cDNA obtained was amplified using degenerate
primers for two conserved regions of Arabidopsis
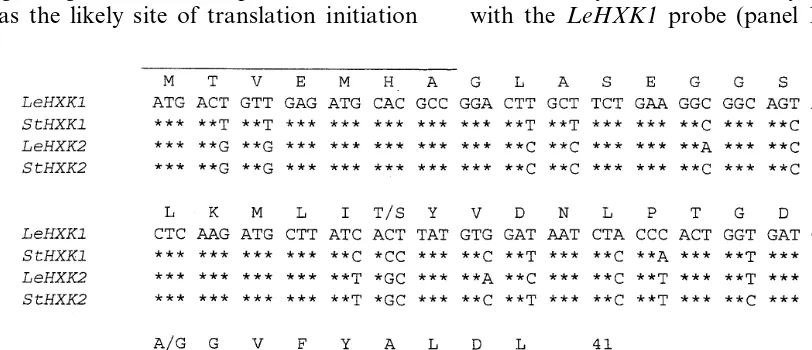
and potato HXKs. A single band about 700 bp long was obtained, purified and subcloned. Several clones were partially sequenced and hinted at the presence of two different HXK genes (Fig. 1). Of the 125 nucleotides, 17 were different in the two HXK sequences, suggesting considerable diver-gence. However, every nucleotide change but one was in the third position and silent, so that the deduced amino acid sequences were practically identical. The two sequences were designated
LeHXK1 and LeHXK2, according to their higher
similarity to potato StHXK1 or StHXK2,
respec-tively (Fig. 1). Both the sequences were used as probes for screening of a cDNA library prepared from young cherry tomato fruit [10]. A single type
of cDNA clone corresponding to LeHXK2 was
isolated and the clone containing the largest insert
of 1770 bp was sequenced. LeHXK2 contained an
open reading frame of 1491 bp followed by a
142-bp sequence and a poly (A+) tail. The ATG
triplet beginning at nucleotide position 138 was assigned as the likely site of translation initiation
since an in-frame termination codon (TAA) was located at positions 108 – 110. The ORF encoded a putative HXK protein of 496 amino acids with a calculated molecular weight of 53754 D and a pI
of 6.3. The putative LeHXK2 protein showed
sim-ilar homology with the two Arabidopsis HXK
proteins (69% identity), and higher homology with
potato StHXK1 and StHXK2 proteins, 83 and
97% identity, respectively (Fig. 2). ATP and two phosphate binding sites, as well as a sugar recogni-tion site were located in the same highly conserved domains in all of the plant hexokinases (Fig. 2).
3.2. Number of HXK genes in tomato plants
While two different sequences encoding HXK were found by RT-PCR of tomato fruit, only one full-length sequence was recovered by the screen-ing of a fruit cDNA library. To determine the number of different HXK genes in tomato, south-ern blot analyses were carried out on tomato genomic DNA digested with four restriction
en-zymes (Fig. 3). The LeHXK1 and LeHXK2 partial
sequences shown in Fig. 1 were radiolabeled with
32P and hybridized with blots shown in Fig. 3. The
LeHXK2 probe hybridized intensely to a single
band in the digests with EcoRI (E), HindIII (H)
and XbaI (X) (panel A). Other bands hybridized
less intensely. These bands hybridized intensely
with the LeHXK1 probe (panel B and C). These
Fig. 2. Comparison of predicted amino acids ofLeHXK2 with potato HXK (StHXK1,StHXK2) andArabidopsisHXK (AtHXK1,
Fig. 3. Southern hybridization of genomic DNA isolated from leaves of tomato plants. The DNA (10mg) was restricted with
EcoRI (E),HindIII (H), XbaI (X) and BamHI (B), as indi-cated above each lane. The same blot was hybridized succes-sively with the original 700 bpLeHXK2 (A) andLeHXK1 (C) fragments. A duplicate blot was hybridized with theLeHXK1 sequence (B). The position of molecular weight markers (Kbp) are shown at the left of panel (A).
Fig. 4. Growth of DFY632 triple mutant cells transformed with pFL61-LeHXK2 on selective media. LeHXK2 cDNA was subcloned into pFL61 and expressed in DFY632. Yeast cells transformed with pFL61 were used as controls. 110 mM of either Gal, Glc or Fru was added to the selective media (−Ura) for the uracil auxotrophic strain.
higher affinity to Glc. High affinity to Glc was confirmed by determining the kinetic properties of
LeHXK2 in protein extracts from DFY632 yeast
cells with either pFL61 or pFL-LeHXK2,
follow-ing MonoQ chromatography (Fig. 6A). Purified extracts from cells transformed with pFL61 alone had no measurable activity for hexose
phosphory-lation while extracts from cells with pFL-LeHXK2
showed high affinity for Glc and Man, Km of 21
Fig. 5. Substrate specificity of LeHXK2 assayed in a native PAGE. Native PAGE and activity staining were carried out as previously described [15], except that the staining solution included 0.6% agarose. When added, hexoses were present at 1 mM.
results suggest that in tomato, there are two dis-tinct HXK genes, which are sufficiently different to give only slight cross-hybridization.
3.3. Functional identification of LeHXK2 cDNA
To analyze whether LeHXK2 encodes an
au-thentic HXK, LeHXK2 was cloned into a yeast
expression vector pFL61 to obtain pFL-LeHXK2.
pFL-LeHXK2 was expressed in triple mutant yeast cells, DFY632, which are unable to phosphorylate hexoses and therefore are unable to grow on Glc or Fru as carbon sources. As shown in Fig. 4,
DFY632 cells transformed with pFL61-LeHXK2
grew on Glc and Fru while control cells containing only pFL61 failed to grow. These results were retested by native gel analysis of crude extracts prepared from yeast cells with pFL61 or
pFL61-LeHXK2. However, activity was detected with Glc as substrate but not with Fru (Fig. 5). Accord-ingly, the doubling time on Glc was 5 h while that
Fig. 6. Partial purification and characterization of hexokinase from yeast extracts expressing LeHXK2. (A) HPLC-ion ex-change (MonoQ) chromatography of yeast protein extracts separated using a KCl gradient. (B) Hexose phosphorylation activity ofLeHXK2 with Glc, Man and Fru. Note different scales of sugar concentrations. V, nmoles of Glc 6-phosphate, Man 6-phosphate or Fru 6-phosphate produced per mg of protein per min.
Table 2
Effect of HXK inhibitors onLeHXK2a
Activity (%) Concentration
Compound
(mM)
Glucosamine 50 83.896.2 (n=4)
aGlucose phosphorylation activity was measured using
de-salted crude extracts in the presence of 1 mM Glc. The results are expressed as the percentage of activity without inhibitor. N, number of repetitions
carried out with the desalted crude extract
indi-cated that LeHXK2 was weakly inhibited by
man-noheptulose, glucosamine and Glc 6-P but was strongly inhibited by ADP (Table 2).
3.4. Expression of LeHXK2 mRNA
LeHXK2 mRNA was easily detected by
RT-PCR using 2 mg of the total RNA for RT and 670
ng of the product for PCR with specific primers
(S2+AS1). Fig. 7 shows that an approximately
700-bp band was amplified under these conditions in roots, leaves, flowers and developing fruit. These conditions permit the detection of specific mRNAs but not their relative amounts. Modifica-tions were, therefore, introduced in order to per-mit quantitative RT-PCR analyses to be carried
out. Of the 2 mg total RNA used in the oligo-dT
primed RT reaction, aliquots corresponding to 200 ng of the total RNA were amplified in PCR
reactions with primers specific for LeHXK2 and
for Actin. These conditions gave linear
amplifica-and 38 mM, respectively (Fig. 6B and Table 1).
Like most plant HXKs, LeHXK2 had a Km value
for Fru in the millimolar range, 7.7 mM, but a
higherVmaxthan for Glc or Man. Inhibitor studies
Fig. 7. Expression analysis of LeHXK2 in different organs. RT-PCR was carried out with specific primers for LeHXK2 and for Actin as given in Section 2. MG, mature green; Br, breaker.
Table 1
Kinetic parameters ofLeHXK2 enzyme expressed in yeast
Substrate Km (mM) Vmax(nmol/mg) Vmax/Km
Glucose 0.021 158 7.5 0.038
Mannose 142 3.7
Fig. 8. Quantitative RT-PCR of cDNA from different organs and tissues and from different stages of fruit development. RNA (2mg) was used for RT as described in Section 2. 200 ng from the same RT reaction were used as template for quantitative PCR reactions using primers specific forLeHXK2 and Actin (LeAc). PCR products were transferred to nylon membranes and detected by hybridization with specific rabiolabeled probes. Imbibed seeds, seeds placed on moistened filter paper for 5 days at 25°C in the dark.
tion but the products could only be detected by hybridization with radiolabeled probe. Fig. 8
shows that LeHXK2 was most highly expressed in
flowers but had also high expression in ovary tissue at anthesis and in seeds imbibed for 5 days. As quantified with an Instant Imager (Packard),
the induction of LeHXK2 messenger in fruit at
anthesis and in imbibed seeds was 2.5 – 2.7-fold higher than the low-level expression measured in most tissues, while induction in flowers was 292-fold.
4. Discussion
4.1. Is LeHXK2 different from the tomato fruit GLK pre6iously described?
Does LeHXK2 correspond to the GLK activity purified from young tomato fruit by Martinez-Barajas and Randall [9]? The calculated molecular weight of LeHXK2 is the same as that determined for fruit GLK (53 kD) and both enzymes have high affinity for Glc and low activity for Fru.
However, the Vmax/Km ratios show that LeHXK2
has only a slight preference for Glc over Man (2-fold) but fruit GLK has about 20-fold prefer-ence for Glc. In addition, inhibitor studies also appear to differentiate the two enzymes. Like most hexokinases, LeHXK2 was weakly inhibited by glucosamine. Both GLK and LeHXK2 were weakly inhibited with 40 – 50 mM of
mannoheptu-lose and strongly inhibited by ADP, but fruit GLK was strongly inhibited by Glc 6-P at pH 7.0
while LeHXK2 was only weakly inhibited at this
pH. Although the latter difference could be due to failure in post translational modifications as sug-gested to explain the difference in pH responses of a potato HXK cDNA expressed in yeast [6], the kinetic differences in the use of Man lead us to tentatively conclude that LeHXK2 does not corre-spond to fruit GLK.
4.2. LeHXK2 is differentially expressed in de6eloping tomato flower and fruit
In comparison with FRK ([4,5]; Ricard, unpub-lished), HXK expression was more difficult to detect on Northern blots. However, constitutive, albeit low, expression could easily be detected by RT-PCR in all the vegetative organs, as well as in developing fruit. Of the former, leaves consistently showed the lowest expression. Expression was highest in flowers, decreased sharply at anthesis and remained low during fruit development. The RNA labeled ‘flower’ was isolated from the entire fully-developed flower whereas the RNA sample labeled ‘anthesis’ was from ovary tissue at the time of anthesis, separated from petals and sepals. It is,
therefore, likely that the induction of LeHXK2
post-anthesis) but not from leaf or radicle tissues.
High expression levels of LeHXK2 in flowers was
not accompanied by correspondingly increased en-zyme activity (preliminary experiments), suggest-ing a role other than metabolic. Confirmation and identification of this role remain to be done.
4.3. Is LeHXK2 a sugar sensor?
HXK genes were proposed to be sugar sensors
based on results obtained with transgenic Ara
-bidopsis plants overexpressing AtHXK1.
Trans-genic tomato plants overexpressing AtHXK1
exhibited even more dramatic changes in pheno-types such as growth inhibition and accelerated senescence, suggesting that sugar signaling path-ways in tomato might be also mediated by
hexok-inase [1,8]. Furthermore, while transgenic
Arabidopsis plants exhibited mutant phenotypes only in germinating seeds and only in presence of exogenous sugars, mature tomato plants
overex-pressing AtHXK1 exhibited their phenotype
inde-pendent of exogenous sugar [8]. However, potato plants transformed with sense or antisense
con-structs of potato StHXK1 exhibit only minor
phe-notypic changes, and in spite of considerable variations in HXK activity in both leaves and tubers, the overall physiology of the transgenic plants was almost not affected, except for starch accumulation in the leaves of antisense plants [6].
Tomato, potato and tobacco are all Solanacea
species and accordingly LeHXK2 had higher
ho-mology with potato and tobacco HXKs compared with Arabidopsis HXKs, AtHXK1 and AtHXK2. Hence, although conserved binding domains for sugar, phosphate and ATP could be identified and are similarly positioned in all HXKs [1], this
infor-mation affords no insight as to whether LeHXK2
is involved in sugar sensing. Antisense repression of LeHXK2 in transgenic tomato plants is
under-way to allow direct testing of the LeHXK2
in-volvement in sugar sensing similar toAtHXK1 and
AtHXK2.
Acknowledgements
This work was supported by Israel Ministry of Science grant 9440-2-98, by Binational Agricul-tural Research and Development (BARD) grant IS-2894-97 and by Binational Science Foundation
(BSF) grant 97-00250. The authors thank Dr Philippe Raymond for the critical reading of the manuscript.
References
[1] J.-C. Jang, P. Leon, L. Zhou, J. Sheen, Hexokinase as a sugar sensor in higher plants, Plant Cell 9 (1997) 5 – 19. [2] J. Sheen, L. Zhou, J.-C. Jang, Sugars as signaling
molecules, Curr. Opin. Plant Biol. 2 (1999) 410 – 418. [3] K.-D. Entian, J.A. Barnett, Regulation of sugar
utiliza-tion by Saccharomyces cere6isiae, Trends Biol. Sci. 17 (1992) 506 – 510.
[4] Y. Kanayama, N. Dai, D. Granot, M. Petreikov, A. Schaffer, A.B. Bennett, Divergent fructokinase genes are differentially expressed in tomato, Plant Physiol. 113 (1997) 1379 – 1384.
[5] Y. Kanayama, D. Granot, N. Dai, M. Petreikov, A. Schaffer, A. Powell, A.B. Bennett, Tomato fructokinases exhibit differential expression and substrate regulation, Plant Physiol. 117 (1998) 85 – 90.
[6] J. Veramendi, U. Roessner, A. Renz, L. Willmitzer, R.N. Trethewey, Antisense repression of hexokinase 1 leads to an overaccumulation of starch in leaves of transgenic potato plants but not to significant changes in tuber carbohydrate metabolism, Plant Physiol. 121 (1999) 123 – 133.
[7] A. Wiese, F. Groner, U. Sonnewald, H. Deppner, J. Lerchl, U. Hebbeker, U.-I. Flugge, A. Weber, Spinach hexokinase I is located in the outer envelope membrane of plastids, FEBS 461 (1999) 13 – 18.
[8] N. Dai, A. Schaffer, M. Petreikov, Y. Shahak, Y. Giller, K. Ratner, A. Levine, D. Granot, Overexpression of
Arabidopsishexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence, Plant Cell 11 (1999) 1253 – 1266.
[9] E. Martinez-Barajas, D.D. Randall, Purification and characterization of a glucokinase from young tomato (Lycopersicon esculentum L. Mill.) fruit, Planta 205 (1998) 467 – 573.
[10] J. Joube`s, T.-H. Phan, D. Just, C. Rothan, C. Bergou-nioux, P. Raymond, C. Chevalier, Molecular and bio-chemical characterization of the involvement of cyclin-dependent kinase A during the early development of tomato fruit, Plant Physiol. 121 (1999) 857 – 869. [11] M. Minet, M.E. Dufour, F. Lacroute, Complementation
of Saccharomyces cere6isiae auxotrophic mutants by
Arabidopsis thalianacDNAs, The Plant J. 2 (1992) 417 – 422.
[12] R.B. Walsh, D. Clifton, J. Horak, D.G. Fraenkel, Sac
-charomyces cere6isiaenull mutants in glucose
phospho-rylation: metabolism and invertase expression, Genetics 128 (1991) 521 – 527.
[13] H. Ito, Y. Fukada, K. Murata, A. Kimura, Transforma-tion of intact yeast cells treated with alkali caTransforma-tions, J. Bacteriol. 153 (1983) 163 – 168.
[15] M. Bouny, P. Saglio, Glycolytic flux and hexokinase activities in anoxic maize root tips acclimated by hy-poxic pretreatment, Plant Physiol. 111 (1996) 187 – 194. [16] S.L. Dellaporta, J. Wood, J.B. Hicks, A plant DNA
minipreparation, version II, Plant Mol. Biol. Rep. 1 (1983) 19 – 21.
[17] T.C. Verwoerd, B.M.M. Dekker, A. Hoekema, A small-scale procedure for the rapid isolation of plant RNAs, Nucl. Acids Res. 17 (1989) 2362.
[18] A. Moing, C. Rothan, L. Svanella, D. Just, P. Diakou, P. Raymond, J.-P. Gaudillere, R. Monet, Organic acid synthesis during peach fruit development, Physiologia Plantarum 108 (2000) 1 – 10.
[19] P. Bork, C. Sander, A. Valencia, Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galacto-kinase families of sugar galacto-kinases, Prot. Sci. 2 (1993) 31 – 40.