DZULFAQOR
DEPARTMENT OF BIOLOGY
FACULTY OF MATHEMATICS AND NATURAL SCIENCE
BOGOR AGRICULTURAL UNIVERSITY
Apis koschevnikovi in South Kalimantan. Supervised by RIKA RAFFIUDIN and MOCHAMAD CHANDRA W.
Deforestation in Kalimantan forest by logging and conversion to palm oil plantation destroyed many living organism and carbon-rich ecosystems. This condition was a serious threat for native honeybee species from Kalimantan i.e. Apis koschevnikovi (Hymenoptera; Apidae). Recent observations shows that the population of A. koschevnikovi has decreased persistently. Ecological integrity of A. koschevnikovi database need to be preserved, including molecular database. Cytochrome oxidase subunit I (COI) in mitochondrial DNA (mtDNA) can assist by providing suitable molecular marker with DNA barcode-based technology. Currently, there are 15 haplotypes of COI gene of A. koschevnikovi has published in GeneBank database. The objective of this research was to explore the nucleotide variations of A. koschevnikovi in South Kalimantan based on COI gene. Five regencies were observed in South Kalimantan forest; Hulu Sungai Selatan (HSS), Hulu Sungai Tengah (HST), Kotabaru (KB), Tanah Bumbu (TB), and Balangan (BL). Based on alignment of 738 bp of COI A. koschevnikovi, there were 5 haplotypes of nucleotides were found. The common haplotypes found in TB and HST samples, while others specific in other places. None of the samples showed the same sequence with the nucleotide sequences of GeneBank database, hence these five haplotypes were concur to be new haplotypes.
Keywords : DNA barcode, COI, A. koschevnikovi, mitochondrial DNA, haplotype.
ABSTRAK
DZULFAQOR. Variasi Gen Sitokrom Oksidase subunit I (COI) di DNA Mitokondria pada Apis koschevnikovi di Kalimantan Selatan. Dibimbing oleh RIKA RAFFIUDIN dan MOCHAMAD CHANDRA W.
Deforestasi di hutan Kalimantan yang berupa pembalakan dan konversi hutan menjadi perkebunan kelapa sawit menghancurkan habitat organisme dan ekosistem. Kondisi ini merupakan ancaman yang serius terhadap spesies asli yang berasal dari Kalimantan, termasuk Apis koschevnikovi (Hymenoptera; Apidae). Penelitian-penelitian sebelumnya menunjukkan kecendrungan populasi A. koschevnikovi yang terus menurun. Memelihara integritas ekologi A. koschevnikovi sangat diperlukan, termasuk database molekulernya. Sitokrom Oksidase subunit I (COI) di DNA mitokondria (DNAmt) dapat membantu dengan menyediakan penanda molekuler dengan teknologi berbasis DNA barcoding. Saat ini, telah ditemukan sebanyak 15 jenis haplotipe gen COI dari A. koschevnikovi yang telah dipublikasikan di Genbank. Tujuan dari penelitian ini adalah untuk mengetahui variasi genetik dari A. koschevnikovi Kalimantan Selatan berdasarkan gen COI dengan menggunakan metode sekuensing DNA. Contoh yang digunakan dalam penelitian ini berasal dari 5 kabupaten di Kalimantan Selatan; Hulu Sungai Selatan (HSS), Hulu Sungai Tengah (HST), Kotabaru (KB), Tanah Bumbu (TB), and Balangan (BL). Pengurutan nukleotida dari contoh yang digunakan menghasilkan panjang nukleotida sebanyak 738 bp, dan sebanyak 5 haplotipe dari nukleotida telah ditemukan. Haplotipe umum ditemukan dari contoh yang berasal dari Kabupaten HST dan TB. Hasil pengurutan dari contoh ini tidak menunjukkan kesamaan dengan 15 haplotipe dari Genebank, sehingga 5 haplotipe yang ditemukan dalam penelitian ini dapat disimpulkan sebagai haplotipe baru.
DZULFAQOR
Intended to obtain Sarjana Sains
In Department of Biology
DEPARTMENT OF BIOLOGY
FACULTY OF MATHEMATICS AND NATURAL SCIENCE
BOGOR AGRICULTURAL UNIVERSITY
NIM
:
G34061910
Study Program : Biology
Approved by,
Dr. Ir. Rika Raffiudin
Drs. Mochamad Chandra W, M.Sc
Supervisor
Supervisor
Endorsed by
Dr. Ir. Ence Darmo Jaya Supena, M.Si.
Head of Biology Department
finally the minithesis entitled Cytochrome Oxidase subunit I (COI) Gene Variations in Mitochondrial DNA of Apis koschevnikovi in South Kalimantan had been finished. The author would like to send acknowledgements to Dr. Ir. Rika Raffiudin and Drs. Moch. Chandra for their supervision and taught, and also to DIKTI that funded this research.
I am forever grateful to my parents, Lulu, Riska, and my hammies for their love and support. Special thanks to Biology 43 crew for the great friendship, and also to members of Zoology community especially Kak Jesi, Kak Rut, Kak Uche, Ajeng and staffs in Biology Department for their help and support.
Bogor, February 2011
Sumiati.
Page
LIST OF TABLE ...viii
LIST OF FIGURE ...viii
LIST OF APPENDIX ...viii
INTRODUCTION ... 1
Background ... 1
Research Objective ... 1
MATERIALS AND METHODS ... 1
Time and Place ... 1
Honey bee Collection and DNA Extraction ... 1
COI Gene Amplification ... 1
Electrophoresis and DNA Visualization ... 2
Sequencing and DNA Analysis ... 2
RESULT... 3
DNA Amplification ... 3
Sequencing and DNA Analysis ... 3
DISCUSSION ... 9
CONCLUSION ... 10
REFERENCES ... 10
APPENDIX ... 12
LIST OF TABLE
Page 1 Nucleotide primers for amplifying COI gene of A. koschevnikovi and primer positions based on mitochondrial DNA of Apis mellifera (Crozier & Crozier 1993) ... 22 A. koschevnikovi COI gene in published Genebank database (www.ncbi.nlm.nih.gov) ... 3
3 Sample code of A. koschevnikovi used in amplification and sequencing in current study ... 4
4 GC and AT content of A. koschevnikovi COI gene from current studies ... 4
5 Haplotypes variations of A. koschevnikovi from current studies based on COI gene ... 4
substitution method with 1000x bootstrap replication ... 8
LIST OF FIGURE
Page
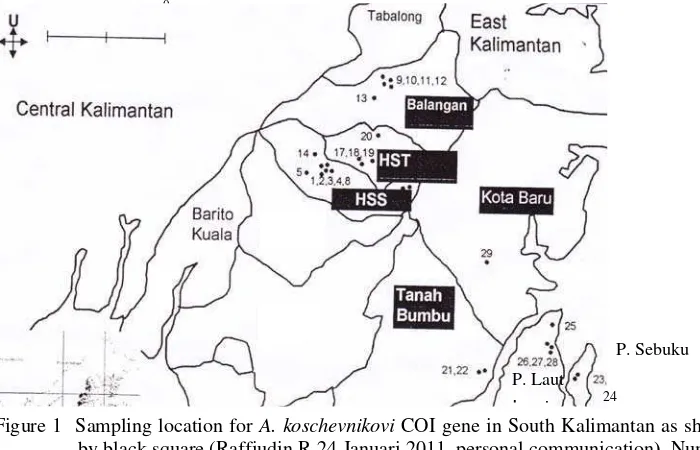
1 Sampling location for A. koschevnikovi COI gene in South Kalimantan as shown by black square
(Raffiudin R 24 Januari 2011, personal communication). Number = colonies number (refer to
Appendix 1).. ... 2
2 DNA fragments of approximately 900 bp COI gene of Apis koschevnikovi. Lane 1-5= Ak 1BL,
Ak 1HST, Ak 8HSS, Ak 1TB, Ak 2KB. M = 100 bp DNA marker (Promega) ... 4
3 Nucleotides sequence of A. koschevnikovi COI region from Ak8 HSS sample and COI putative
amino acids. Number = nucleotide position. Underlined nucleotides = forward and reverse
primer. ... 5
4 Nucleotides alignment of A. koschevnikovi COI gene resulted from this study ... 5
5Putative amino acid sequences of A. koschevnikovi COI gene in this study. Number = amino
acid position, = variation of amino acid based on six samples in this study, : = closely related
of amino acid ... 6
6 Phylogenetic tree of A. koschevnikovi from current studies and 15 haplotypes in Genebank
databases based on COI region using NJ construction with 1000x bootstrap replication.. ... 9
LIST OF APPENDIX
Page
1 A. koschevnikovi honey bee collection from South Kalimantan. * = samples used for sequencing
and analysing in this study (Raffiudin R 24 Januari 2011, personal communication) ... 13
2 Chromatogram of A. koschevnikovi cytochrome oxidase subunit 1 (COI) gene from Ak8 HSS
sample using AmF_cox1 forward primer ... 14
3 Chromatogram of A. koschevnikovi cytochrome oxidase subunit 1 (COI) gene from Ak 8HSS
INTRODUCTION
Background
Deforestation in Kalimantan due to logging and forest conversion to palm oil plantation destroyed many living organism and carbon-rich ecosystems (Curran et al.
2004). This condition was a serious threat for the species that is native in Kalimantan such as Apis koschevnikovi,a cavity nesting honey bee. It is classified in the order of Hymenoptera, suborder of Apocrita, superfamily of Apoidea, and family of Apidae (Michener 2000). The distribution of A. koschevnikovi has establishes many new site localities but all within the previously confirmed limits of the tropical evergreen forest of Sundaland (Hadisoesilo et al. 2008). Nowadays, recent observations shows that the population of A. koschevnikovi has decreased persistently due to forest loss and establishment of some clear land to make oil palm, rubber, and coconut plantations (Otis 1991).
Preserving the ecological integrity of A. koschevnikovi database is necessary, including molecular database. Cytochrome oxidase subunit I (COI) in mitochondrial DNA (mtDNA) provide suitable molecular marker with DNA barcode-based technology due to their characteristic i.e no deletion and insertion in its sequence, and slightly variances in their sequences (Hebert et al.
2003). In animals mtDNA, COI usually composed by over 600 bp of long, and at least 648 bp of long is required to be submitted into Genebank database (www.ncbi.nlm.nih.gov).
Currently, there are 15 haplotypes of COI gene of A. koschevnikovi has published in Genebank under Accesion number AF153110 - AF153111, AY012723, AY754729 - AY754732, and DQ016097 - DQ016104 (www.ncbi.nlm.nih.gov). Most of the haplotypes occured from Sabah (haplotype 3-7, hap 11-15), the others are from Sarawak (Malaysia), and West Kalimantan. However, out of 15 haplotypes, three found in Kalimantan. This study was aimed to explore further to find another variations of COI haploytpes for building the database especially in Kalimantan forest.
Research Objective
The objective of this research was to explore the variations of COI gene in
mitochondrial DNA of A. koschevnikovi in South Kalimantan.
MATERIALS AND METHODS
Time and Place
The research was conducted on January 2010 up to February 2011, and it was taken place in Division of Animal Function and Behaviour, Department of Biology, Bogor Agricultural University (IPB).
Honey Bee Collection and DNA Extraction
The objects of my research were DNA from several A. koschevnikovi honey bees. Honey bees used in this study were collection of Dr. Rika Raffiudin (Dept. Biology IPB) that were collected from Kabupaten Hulu Sungai Selatan (Ak HSS), Hulu Sungai Tengah (Ak HST), Balangan (Ak BL), Tanah Bumbu (Ak TB), and Kotabaru (Ak KB) in South Kalimantan (Figure 1). Several worker honey bee from several colonies were collected from each regencies and were preserved in absolute ethanol.
DNA extraction has carried out by using 0.2% Cetyl Trimethyl Ammonium Bromide (CTAB) and ethanol precipitation (Sambrook
et al. 1989).
COI Gene Amplification
P. Sebuku minutes (mins) at 940 C for initial denaturing, 30
Table 1 Nucleotide primers for amplifying COI gene of A. koschevnikovi and primer positions based on mitochondrial DNA of Apis mellifera (Crozier & Crozier 1993)
minutes (mins) at 940 C for initial denaturing, 30 cycles of 1 min at 940 C (denaturing), 1 min and 30 seconds at 550 C (annealing), 2 mins at 720 C (DNA elongation) and followed by 5 mins for final extension.
Electrophoresis and DNA Visualization
Amplified DNA was migrated using 6% Polyacrilamid Gel Electrophoresis (PAGE) which was submerged in 1x TBE buffer (10 mM Tris-HCl, 1 M Borat acid, and 0.1 mM EDTA), operated for 50 minutes at 200 V. The compositions of 6% gel acrilamide were 12 ml of aquades, 4 ml of 5xTBE, 4 ml of acrilamide, 15 µl of TEMED, and 160 µl of 10% APS.
DNA visualization used sensitive Silver Nitrate staining (Tegelstrom 1986). Gel resulting from electrophoresis was washed using CTAB solution (0.2 g/200 ml of aquades) for 8 mins, and was flushed using 200 ml of aquades for 2 mins (2 times). Subsequently, its gel was submerged in NH4OH solution (2.4 ml/200 ml of aquades)
for 8 mins, and solution of mixture contained 0.32 g of AgNO3, 0.8 ml of NH4OH, 80 µl
NaOH, and 200 ml of aquades for 10 mins. Then, the gel was flushed again using the same condition as above, and was submerged again in the solution of mixture contained 4 g of NA2CO3, 100 µl of formaldehida, and 200
ml of aquades until the bands of DNA seen. Staining process was stopped using 200 µl of 1% acetat acid in 200 ml of aquades.
Sequencing and DNA Analysis
Genetic variations in COI mitochondrial region was analysed based on DNA sequencing method. DNA was sequenced by using ABI 3100 and 3730XL sequencer machine at sequencing company. Nucleotide sequence were edited and analysed using Genetyx Win Version 4.0 software. Forward and reverse sequence were aligned to obtain contiguous sequence by using Clustal X software (Thompson et al. 1997). All DNA contiguous sequences was aligned together with haplotypes databases of COI gene of A. koschevnikovi in Genebank NCBI (National Center for Biotechnology Information) under Accesion number AF153110-AF153111, Primer Nucleotide sequence of primer
(5’-3’) (Crozier & Crozier 1993)
Positions primer in mitochondrial DNA of Apis mellifera
(Crozier & Crozier 1993) Forward amF_cox1 CCCCAGGATCATGAATTAGC 1915-1934 Reverse amR_cox1 TCATTGCTGTACCAACAGGAA 2719-2739 Figure 1 Sampling location for A. koschevnikovi COI gene in South Kalimantan as shown
by black square (Raffiudin R 24 Januari 2011, personal communication). Number = colonies number (refer to Appendix 1).
P. Laut
AY012723, AY754729- AY754732, and DQ016097-DQ016104
(www.ncbi.nlm.nih.gov) (Table 2).
Edited DNA sequences were translated into putative amino acid using Genetyx and were aligned by using Clustal X software.
DNA genetic distances and phylogenetic analysis were constructed between current samples and 15 haplotypes in Genebank as ingroup while A. cerana with accesion number GQ162109, and A. mellifera with accesion number L06178 as outgroups. These analysis was carried out by using MEGA4 software based on Kimura-2-parameter substitution method and Neighbour Joining
(NJ) construction with 1000x bootstrap (Tamura et al. 2007).
RESULT
DNA Amplification
There were 29 samples of A. koschevnikovi
COI gene from five regencies in South Kalimantan were amplified using AmF_cox1 and AmR_cox1 primer (Table 3). All samples showed the same results i.esingle band with approximately 900 bp of nucleotide (Figure 2).
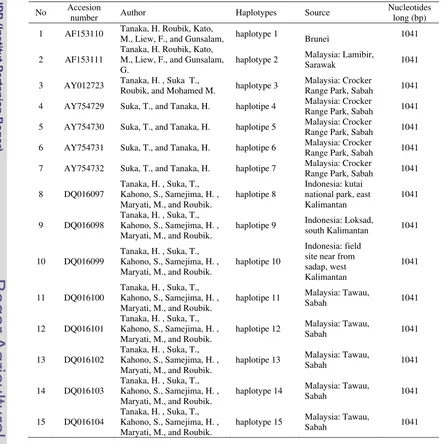
Table 2 A. koschevnikovi COI gene in published Genebank database (www.ncbi.nlm.nih.gov)
No Accesion
number Author Haplotypes Source
Nucleotides long (bp)
1 AF153110 Tanaka, H. Roubik, Kato,
M., Liew, F., and Gunsalam, haplotype 1 Brunei 1041
2 AF153111
Tanaka, H. Roubik, Kato, M., Liew, F., and Gunsalam, G.
haplotype 2 Malaysia: Lamibir,
Sarawak 1041
3 AY012723 Tanaka, H. , Suka T.,
Roubik, and Mohamed M. haplotype 3
Malaysia: Crocker
Range Park, Sabah 1041
4 AY754729 Suka, T., and Tanaka, H. haplotipe 4 Malaysia: Crocker
Range Park, Sabah 1041
5 AY754730 Suka, T., and Tanaka, H. haplotipe 5 Malaysia: Crocker
Range Park, Sabah 1041
6 AY754731 Suka, T., and Tanaka, H. haplotipe 6 Malaysia: Crocker
Range Park, Sabah 1041
7 AY754732 Suka, T., and Tanaka, H. haplotipe 7 Malaysia: Crocker
Range Park, Sabah 1041
8 DQ016097
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 8
Indonesia: kutai national park, east Kalimantan
1041
9 DQ016098
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 9 Indonesia: Loksad,
south Kalimantan 1041
10 DQ016099
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 10
Indonesia: field site near from sadap, west Kalimantan
1041
11 DQ016100
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 11 Malaysia: Tawau,
Sabah 1041
12 DQ016101
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 12 Malaysia: Tawau,
Sabah 1041
13 DQ016102
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 13 Malaysia: Tawau,
Sabah 1041
14 DQ016103
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotype 14 Malaysia: Tawau,
Sabah 1041
15 DQ016104
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotype 15 Malaysia: Tawau,
Sequencing and DNA Analysis
Out of 29 amplified samples of A. koschevnikovi COI gene, six samples were sequenced (see Table 3 and Appendix 1). Nucleotide sequence of AmR_cox1 primer was found in AmF_cox1 sequence (Appendix
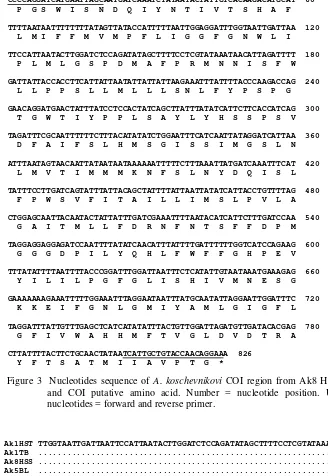
2), while nucleotide sequence of AmF_cox1 primer found in AmR_cox1 sequence (Appendix 3). Complete contiguous sequence was found in Ak HSS sample colony number 8 (Ak8 HSS) i.e 826 bp (Figure 3).
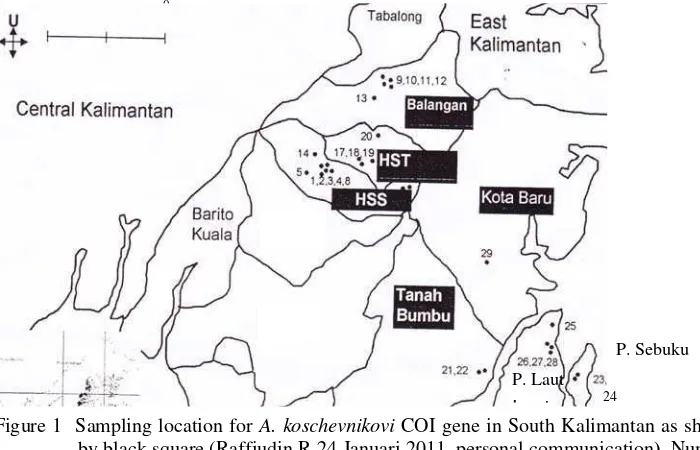
Based on six contiguous of A. koschevnikovi COI gene sequences alignment, 738 bp of nucleotide was observed. All contiguous DNA showed rich of A-T sequences, i.e. 75.5 % while G-C was 24.5 % (Table 4).
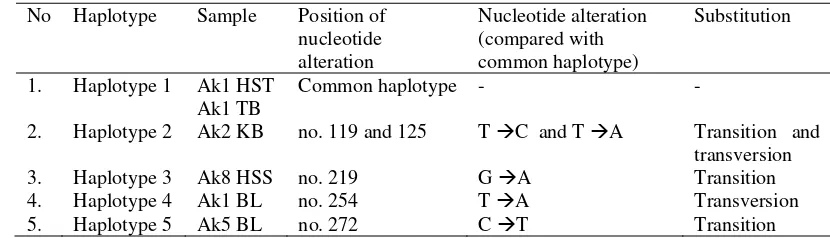
There were five variations of DNA sequences occurred from current studies alignment. The variations respectively occurred in nucleotide number 119, 125, 219, 254 and 272 (Figure 4). Hence, five haplotypes were observed (Table 5). The first haplotype was a common haplotypes was found in Ak1 TB and Ak1 HST samples. The second to fifth haplotypes respectively were found in Ak2 KB, Ak8 HSS, Ak1 BL, and Ak5 BL sample (Table 5).
Table 3 Sample code of A. koschevnikovi used in amplification and sequencing in current study
Table 4 GC and AT content of A. koschevnikovi COI gene from current studies
No Sample Code A-T nucleotides (%) G-C nucleotides (%)
1 Ak1 HST 75.53 24.47
2 Ak1 TB 75.53 24.47
3 Ak8 HSS 75.67 24.33
4 Ak5 BL 75.67 24.33
5 Ak1 BL 75.53 24.47
6 Ak2 KB 75.39 24.61
Table 5 Haplotypes variations of A. koschevnikovi from current studies based on COI gene
No Haplotype Sample Position of nucleotide alteration
Nucleotide alteration (compared with common haplotype)
Substitution
1. Haplotype 1 Ak1 HST Common haplotype - -
Ak1 TB
2. Haplotype 2 Ak2 KB no. 119 and 125 T ÆC and T ÆA Transition and transversion
3. Haplotype 3 Ak8 HSS no. 219 G ÆA Transition
4. Haplotype 4 Ak1 BL no. 254 T ÆA Transversion
5. Haplotype 5 Ak5 BL no. 272 C ÆT Transition
No Sample code
Regency Origin Number of amplified colonies
Samples for sequenced and alignment analysis
1 Ak HSS Hulu Sungai Selatan 11 Colony no.8 (Ak8 HSS) 2 Ak HST Hulu Sungai Tengah 4 Colony no.1 (Ak1 HST) 3 Ak BL Balangan 5 Colony no.1 & 5 (Ak1 BL &
Ak5 BL)
4 Ak KB Kotabaru 7 Colony no. 2 (Ak2 KB)
5 Ak TB Tanah Bumbu 2 Colony no. 1 (Ak1 TB)
1 2 3 4 5 M
900 bp
400 bp
Figure 3 Nucleotides sequence of A. koschevnikovi COI region from Ak8 HSS sample and COI putative amino acid. Number = nucleotide position. Underlined nucleotides = forward and reverse primer.
CCCCAGGATCATGAATTAGCAATGATCAAATCTATAATACTATTGTCACAAGACATGCAT 60 P G S W I S N D Q I Y N T I V T S H A F
TTTTAATAATTTTTTTTATAGTTATACCATTTTTAATTGGAGGATTTGGTAATTGATTAA 120 L M I F F M V M P F L I G G F G N W L I
TTCCATTAATACTTGGATCTCCAGATATAGCTTTTCCTCGTATAAATAACATTAGATTTT 180 P L M L G S P D M A F P R M N N I S F W
GATTATTACCACCTTCATTATTAATATTATTATTAAGAAATTTATTTTACCCAAGACCAG 240 L L P P S L L M L L L S N L F Y P S P G
GAACAGGATGAACTATTTATCCTCCACTATCAGCTTATTTATATCATTCTTCACCATCAG 300 T G W T I Y P P L S A Y L Y H S S P S V
TAGATTTCGCAATTTTTTCTTTACATATATCTGGAATTTCATCAATTATAGGATCATTAA 360 D F A I F S L H M S G I S S I M G S L N
ATTTAATAGTAACAATTATAATAATAAAAAATTTTTCTTTAAATTATGATCAAATTTCAT 420 L M V T I M M M K N F S L N Y D Q I S L
TATTTCCTTGATCAGTATTTATTACAGCTATTTTATTAATTATATCATTACCTGTTTTAG 480 F P W S V F I T A I L L I M S L P V L A
CTGGAGCAATTACAATACTATTATTTGATCGAAATTTTAATACATCATTCTTTGATCCAA 540 G A I T M L L F D R N F N T S F F D P M
TAGGAGGAGGAGATCCAATTTTATATCAACATTTATTTTGATTTTTTGGTCATCCAGAAG 600 G G G D P I L Y Q H L F W F F G H P E V
TTTATATTTTAATTTTACCCGGATTTGGATTAATTTCTCATATTGTAATAAATGAAAGAG 660 Y I L I L P G F G L I S H I V M N E S G
GAAAAAAAGAAATTTTTGGAAATTTAGGAATAATTTATGCAATATTAGGAATTGGATTTC 720 K K E I F G N L G M I Y A M L G I G F L
TAGGATTTATTGTTTGAGCTCATCATATATTTACTGTTGGATTAGATGTTGATACACGAG 780 G F I V W A H H M F T V G L D V D T R A
CTTATTTTACTTCTGCAACTATAATCATTGCTGTACCAACAGGAAA 826 Y F T S A T M I I A V P T G *
Ak1HST TTGGTAATTGATTAATTCCATTAATACTTGGATCTCCAGATATAGCTTTTCCTCGTATAAATAACATTA [138]
Ak1TB ... [138]
Ak8HSS ... [138]
Ak5BL ... [138]
Ak1BL ... [138]
Ak2KB ...C...A... [138]
Ak1HST CAGGATGAACTGTTTATCCTCCACTATCAGCTTATTTATATCATTCTTCACCATCAGTAGATTTCGCAA [276] Ak1TB ... [276]
Ak8HSS ...A... [276]
Ak5BL ...T.... [276]
Ak1BL ...A... [276]
Ak2KB ... [276]
Ak2KB FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 Ak8HSS FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 Ak1BL FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 Ak1HST FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 Ak1TB FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 Ak5BL FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 AmCrozier FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLFMLLLSNLFYPSPGTG 60 AkHap13 FLIGGFGNWLIPLMLGSPDMAFPRMNNISFWLLPPSLLMLLLSNLFYPSPGTG 60 *************************************:***************
Ak2KB GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 Ak8HSS GTGWTIYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 Ak1BL GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 Ak1HST GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 Ak1TB GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 Ak5BL GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 AmCrozier GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 AkHap13 GTGWTVYPPLSAYLYHSSPSVDFAIFSLHMSGISSIMGSLNLMVTIMMMKNFS 120 *****:***********************************************
Figure 5 Putative amino acid sequence of A. koschevnikovi COI gene from this study. Number = amino acid position, = variation of amino acid based on six samples in this study, : = closely related of amino acid.
Nucleotide sequence of Ak8 HSS were translated into putative amino acid and showed no stop codon (Figure 3). Putative amino acid of COI gene from current studies were aligned as well. The result showed that there was one amino acidvariation sequence (Figure 4) i.e at nucleotide no. 219-221, codon
ATTwas translated into Isoleusin (I) in Ak8 HSS sample (haplotype 3), while Valin (V) was resulted from codon GTT in Ak1 HST and Ak1 TB sequences (common haplotype).
Meanwhile, the other variations from current studies alignment did not show variations for its putative amino acid. For examples, In Ak2 KB sample, codon TTC at nucleotides no. 117-119 (haplotype 2) was translated into Phenilalanin (F) and codon
CGAat nucleotides no. 123-125 was translated into Arginin (R). At the same position in common haplotype, codon TTTand CGT were translated become same amino acid, Phenilalanin and Arginin (Figure 5). In Ak1 BL sample, codon TCA at nucleotides no. 252-254 (haplotype 4) was translated become Serin (S). At the same position in common haplotypes, codon TCT was translated become Serin amino acid as well. The other example came from Ak5 BL sample, codon TTT at nucleotides no. 270-272 (haplotype 5) was translated into Phenilalanin (F). At the same position in common haplotypes, codon TTC
was translated become Phenilalanin amino acid as well.
This study performed alignment between current sequenced samples (refer to Table 3) and published A. koscehvnikovi COI databases in Genebank as well. Total of 685 bp of nucleotides was aligned. None of the current study COI A. koschevnikovi study samples showed the same sequence with the published nucleotide sequences from Genebank databases (Table 6).
Table 6 Position of nucleotide variations of A, koschevnikovi samples in COI gene. Common hap= Ak1 HST and Ak1 TB. Source of hap1-hap15 refer to Table 2
1 1 2 3 4 4 5 5 5 6 6 7 8 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 4 4 4 4 2 5 1 3 3 5 1 4 7 3 6 2 1 2 2 3 4 4 5 5 5 5 6 6 7 7 0 0 1 3 8 0 2 4 7 8 9 0 0 1 3 4 4 5 5 6
Hap 4 6 8 1 7 0 3 6 9 5 6 7 8 1 7 9 1 2 9 1 5 2 7 3 5 9 2 5 1 7 0 9 2
Common hap A A T T C T T A T T T T C T A T C A A A A A T G A C T A C T A T T A T T T A C T A C T A A A
Ak2 KB . . . . . . . . C A . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
Ak8 HSS . . . . . . . . . . . . . . . . . . . . . A . . . . . . . . . . . . . . . . . . . . . .
Ak1 BL . . . . . . . . . . . . . . . . . . . . . . . . A . . . . . . . . . . . . . . . . . . .
Ak5 BL . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . . . . . . . . . . . .
Hap10_Kalbar . . . C . . . . . A C A . . . . T . . . . . . . . . . . . C . . . . . . . . T . . . C T . T
Hap11_Sabah . . . C . . . . . A C A . . . . T . . . . . . . . . . . . C . . . . . . . T T . . . C T . T
Hap2_Sarawak . . . C . . . . C A C A . . . . T . . . . . . . . T . . . C . . . . . . . . T . . . C T . T
Hap14_Sabah . . . . . . . A . . . . . . . G . . . . . . . . . . . . . C . . . . . . T . . . . . . .
Hap15_Sabah . . . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . .
Hap9_Kalsel . . . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . .
Hap12_Sabah . . . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . .
Hap1_Brunei . . . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . G . . . . .
Hap6_Sabah . . . . . . . A . . . . . . . . . . . . . . C . . . . . . . . . . . . . T . . . . . . .
Hap7_Sabah . . . . . G . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . .
Hap13_Sabah . . . . . . . A . . . . . . . . . . . . . . . . . . T . . . . . . . . . T . . . . . . .
Hap8_Kaltim . G . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . . . . .
Hap5_Sabah . . . . . . . A . . . . . . . . . . . . . . . . . . . . . . . . . . . . T . . . C . . .
Hap3_Sabah T . A . T A A . . A . . T A T C T . T T T T A . . . A T T A T . C T C A A T . C . T C . T .
Hap4_Sabah T . A . T A A . . A . . T A T C T . T T T T A . . . A T T A T . C T C A A T . C . T C . T .
4 4 4 5 5 5 5 5 6 6 6 6 6 6 6 6 6 6 6 7 3 3 4 7 9 0 2 3 3 4 5 6 7 8
Hap 5 8 7 1 4 0 3 1 0 7 1 9 5 1 9 8 5
Common hap T A T T A A A A A A C T T T T T A
Ak2 KB . . . . . . . . . . . . . . .
Ak8 HSS . . . . . . . . . . . . . . .
Ak1 BL . . . . . . . . . . . . . . .
Ak5 BL . . . . . . . . . . . . . . .
Hap10_Kalbar C . . . . . . . T . . . . . .
Hap11_Sabah C . . . . . . . T . . . . . .
Hap2_Sarawak . . . . . . . . T . . . . . .
Hap14_Sabah . . . . G . . . . . . . . . . . .
Hap15_Sabah . . . . . . . G . . . . . . .
Hap9_Kalsel . . . . . . . . . . . . . . .
Hap12_Sabah . . . . . . . . . . . . . . .
Hap1_Brunei . . . . . . . . . . . . . . .
Hap6_Sabah . . . T . . . . . . . . . . . . .
Hap7_Sabah . . . A . . . . . . . . . . . . .
Hap13_Sabah . . . . . . . . . . . . . . .
Hap8_Kaltim . . . . . . . . T . . . . . .
Hap5_Sabah . . C . . . . . . . . . . . . . .
Hap3_Sabah C T C T . T T T T . T C A A . C .
Table 7 Genetic distances of A. koschevnikovi COI gene (current study, refer to Table 3) and 15 haplotypes of COI gene from Genebank (refer to Table 2) based on Kimura-2-parameter substitution method with 1000x bootstrap
[ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 ]
[ 1] [ 2] 0.001 [ 3] 0.070 0.072 [ 4] 0.070 0.072 0.000 [ 5] 0.073 0.075 0.006 0.006 [ 6] 0.071 0.073 0.015 0.015 0.018 [ 7] 0.075 0.077 0.018 0.018 0.021 0.003 [ 8] 0.071 0.073 0.015 0.015 0.018 0.000 0.003 [ 9] 0.075 0.077 0.018 0.018 0.021 0.003 0.006 0.003 [10] 0.073 0.075 0.016 0.016 0.019 0.001 0.004 0.001 0.004 [11] 0.072 0.073 0.018 0.018 0.021 0.003 0.006 0.003 0.006 0.004 [12] 0.073 0.075 0.018 0.018 0.021 0.003 0.006 0.003 0.006 0.004 0.004 [13] 0.071 0.073 0.015 0.015 0.018 0.003 0.006 0.003 0.006 0.004 0.006 0.006 [14] 0.068 0.070 0.015 0.015 0.018 0.003 0.006 0.003 0.006 0.004 0.006 0.006 0.006 [15] 0.070 0.072 0.016 0.016 0.019 0.001 0.004 0.001 0.004 0.003 0.004 0.004 0.004 0.004 [16] 0.072 0.073 0.018 0.018 0.021 0.003 0.006 0.003 0.006 0.004 0.006 0.006 0.006 0.006 0.004 [17] 0.072 0.073 0.018 0.018 0.021 0.003 0.006 0.003 0.006 0.004 0.006 0.006 0.006 0.006 0.004 0.000 [18] 0.073 0.075 0.019 0.019 0.022 0.004 0.007 0.004 0.007 0.006 0.007 0.007 0.007 0.007 0.006 0.001 0.001 [19] 0.070 0.072 0.019 0.019 0.022 0.004 0.007 0.004 0.007 0.006 0.007 0.007 0.007 0.007 0.006 0.001 0.001 0.003 [20] 0.070 0.072 0.019 0.019 0.022 0.004 0.007 0.004 0.007 0.006 0.007 0.007 0.007 0.007 0.003 0.001 0.001 0.003 0.003 [21] 0.071 0.073 0.015 0.015 0.018 0.006 0.009 0.006 0.009 0.007 0.009 0.009 0.009 0.009 0.007 0.003 0.003 0.004 0.004 0.004 [22] 0.109 0.107 0.092 0.092 0.092 0.088 0.092 0.088 0.090 0.088 0.090 0.088 0.087 0.092 0.087 0.092 0.092 0.094 0.090 0.090 0.092 [23] 0.102 0.102 0.096 0.096 0.092 0.093 0.096 0.093 0.096 0.094 0.093 0.093 0.096 0.096 0.091 0.093 0.093 0.095 0.095 0.091 0.089 0.112
[ 1] #Ak_T_hap3_Sabah [ 6] #Ak_T_hap9_Kalsel [11] #Ak_T_hap6_Sabah [ 16] #Ak1HST_R_Kalsel [ 21] #Ak2KB_R_KAlsel
[ 2] #Ak_T_hap4_Sabah [ 7] #Ak_T_hap15_Sabah [12] #Ak_T_hap7_Sabah [ 17] #Ak1TB_R_Kalsel [ 22] #Ac_china
[ 3] #Ak_T_hap10_Kalbar [ 8] #Ak_T_hap12_Sabah [ 13] #Ak_T_hap8_Kaltim [ 18] #Ak8HSS_R_Kalsel [ 23] #Am_Crozier
[ 4] #Ak_T_hap11_Sabah [ 9] #Ak_T_hap14_Sabah [ 14] #Ak_T_hap5_Sabah [ 19] #Ak1BL_R_Kalsel
Ak T hap15 Sab ah
Ak T hap12 Sab ah
Ak T hap14 Sab ah
Ak T hap1 Brunei
Ak T hap9 Kalsel
Ak T hap6 Sab ah
Ak T hap7 Sab ah
Ak T hap8 Kaltim
Ak T hap5 Sab ah
Ak T hap13 Sab ah
Ak 5BL R Kalsel
Ak 1BL R Kalsel
Ak 1HST R Kalsel
Ak 1TB R Kalsel
Ak 8HSS R Kalsel
Ak 2KB R KAlsel
Ak T hap2 Sarawak
Ak T hap10 Kalb ar
Ak T hap11 Sab ah
Ak T hap3 Sab ah
Ak T hap4 Sab ah
Apis cerana
Apis m ellifera 100 66 92 98 100 88 46 49 37 34 65 28 23 19 26
0.01
Figure 6 Phylogenetic tree of A. koschevnikovi from current studies and 15 haplotypes in Genebank databases based on COI region using NJ method with 1000x bootstrap. Samples code refer to Table 2 and 3.
DISCUSSION
The result of A. koschevnikovi COI gene amplification in PAGE 6% showed approximately about 900 bp of nucleotides long, while 801 bp of average nucleotides long resulted from DNA sequencing toward current samples of A. koschevnikovi. This difference might be occur due to gel shifting phenomenon i.e. the alteration of DNA band movement in polyacrilamide gel.
DNA sequencing result showed different size of nucleotides for each sample. This condition was due to the unclear graph in beginning of chromatogram. For alignment purpose, all contiguous sequences must commence and end at the same nucleotide position.
The high number of AT nucleotides i.e 75.5% from each samples agree with insect DNAmt that contained rich of A+T base (Crozier et al. 1989). Longer time was needed to amplify DNA which has a very high G-C content (Griffiths et al. 1999).
There were two kind of nucleotide substitution occurred from current studies alignment i.e transition and transversion
(Table 5). Transition occurred when nucleotide substitution did not change the structure of nitrogene base i.e purin (A and G) to be purin; or pirimidin (C and T) to be pirimidin, while transversion occured when nucleotide substitution change the structure of nitrogene base (purin to be pirimidin; or pirimidin to be purin) (Griffiths et al. 1999).
The nucleotide substitution does not always resulted amino acid from transcription-translation process. The alteration will effect translated amino acid when the substitution occurred in first or second nucleotide in codon formation (Klug et al. 2006). For the example in Ak8 HSS sample at nucleotide no. 219-221, codon ATTwas translated into Isoleusin (I) in Ak8 HSS (haplotype 3), while Valin (V) was resulted from codon GTT from Ak1 HST and Ak1 TB sequence (common haplotype). However, Valin and Isoleusin is still closely related in amino acid as showed with double dot (:) (see Figure 5). Both of them were clustered in hydrophobic aliphatic amino acid (Takei et al. 2006).
Cluster A
Cluster B
In addition, DNA variation especially third nucleotide substitution in codon formation does not always produce different putative protein (Avise 1994). The examples based on Ak2 KB sample (haplotype 2) at nucleotides no. 117-119 and no. 123-125, Ak1 BLsample at nucleotides no. 252-254 (haplotype 4), and Ak5 BL sample at nucleotides no. 270-272 (haplotype 5).
The alignment between current samples (refer to Table 3) and published A. koschevnikovi COI databases in Genebank has resulting 685 bp of nucleotides long (Table 5), which fulfilled 648 nucleotides as a minimum number of nucleotide requirement to be submitted to DNA Barcode (www.ncbi.nlm.nih.gov). None of the current COI A. koschevnikovi study samples showed the same sequence with the published nucleotide sequence from Genebank databases (Table 5). Hence, these five haplotypes from this research were the new haplotypes for A. koschevnikovi COI database.
The results of this research can be used to accomplish information about molecular ecology studies of A. koschevnikovi as well. Based on current studies alignment, Ak1 HST and Ak1 TB has a same sequence, although the distance of HST and TB regencies approximately 150 km(Figure 1). However, sequences variations were found in two samples both from Balangan Regency (Ak1 BL and Ak5 BL). These results indicate that mixed population occurred in A. koschevnikovi distribution.
Genetic distances and phylogenetic analysis were performed to observe genetic relationship between current studies and published A. koschevnikovi COI databases in Genebank as well. Genetic distance analysis grouped all samples in this study in one cluster, with the highest genetic distance in Ak2 KB sample i.e 0.003-0.004 (Table 7). Ak2 KB sample was collected not from Kalimantan mainland but from Sebuku island (see sampling location number 24 in Figure 1). Hence, these highest number might be due to geographical barrier which separate by Laut strait and Laut island (see Figure 1).
Phylogenetic analysis grouped Ak samples from this previous study in three clusters (Figure 6). All samples in this study and several haplotypes from Sabah and Brunei were clustered in cluster A (0.000-0.009 value of genetic distances), while haplotype 2 from Sarawak, haplotype 10 from West Kalimantan, and haplotype 11 from Sabah were clustered in cluster B (0.000-0.022 value
of genetic distances) (Table 7). However, both were classified in one group due to their bootstrap value which is 100 (Figure 6).
However, Ak haplotypes 3 and 4 from Sabah were grouped in different cluster with 100 of bootstrap value, while these samples were collected from same region with several haplotypes in cluster A and B. Genetic distances of haplotypes 3 and 4 compared with the other haplotypes showed the highest value (0.001-0.077) (Table 7). Therefore, these 2 haplotypes need to be reconfirmed for the species validity.
In the case of these two anomalous haplotypes from Genebank, therefore, one need to be aware with the published DNA database. In addition, further studies from the other areas in Kalimantan is necessary to have a complete picture of A. koschevnikovi COI gene database.
CONCLUSION
The exploration of COI gene of A. koschevnikovi from six samples taken from five regencies in South Kalimantan showed the genetic differentation. There were five haplotypes of A. koschevnikovi COI gene were found in this study i.e. haplotype 1, 2, 3, 4, and 5. Haplotype 1 was a common haplotype was found in sample Ak1 HST and Ak1 TB, while others specific in other places. None of the current samples showed the same sequence with the nucleotide sequences of Genebank database, hence these five haplotypes were the new haplotypes of A. koschevnikovi COI gene.
REFERENCES
Avise JC. 1994. Molecular Markers, Natural History and Evolution. New York: Chapman & Hall.
Crozier RH, Crozier YC, Mackinlay AG. 1989. The CO-I and CO-II region of honeybee mitochondrial DNA: evidence for variation in insect mitochondrial evolutionary rates.
Mol Biol Evol 6: 399-411
Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization,
Genetics 133: 97-117.
Griffiths A, William MG, Jeffrey HM, Richard CL. 1999. Modern Genetic Analysis. New York: W.H. Freeman and Company.
Hadisoesilo et al. 2008. Morphometric analysis and biogeography of Apis koschevnikovi Enderlein (1906),
Apidologie 39: 495-503.
Hebert PD, Ratnasingham S, dewaard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species, Proc. R. Soc. Lond 270: S96-S99.
Klug WS, Cummings MR, Spencer CA. 2006.
Concepts of Genetics 8th ed. New Jersey: Pearson Education, Inc. Michener CD. 2000. The Bees of the World.
London: The John Hopkins Univ Pr. Otis GW. 1991. A review of the diversity of
species within Apis. In: Smith DR (ed) Diversity in The Genus Apis.
Oxford: Westview Press. pp 29-50. Sambrook J, Fritsch EF, Maniatis T. 1989.
Molecular Cloning: A Laboratorium Manual Second Edition. New York:
Cold Spring Harbor Laboratory Press.
Takei T et al. 2006. The effects of the side chains of hydrophobic aliphatic amino acid residues in an amphipathic polypeptide on the formation of a helix and its association, J. Biochem 139: 271-278.
Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0, Mol Biol Evol 24: 1596-1599.
Tegelstrom H. 1986. Mitochondrial DNA in natural population: an improved routine for screening of genetic variation based on sensitive silver staining. Electrophoresis 7: 226-229. Thompson JD et al. 1997. The clustal X
windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acid Res 24: 4876-4882.
Appendix 1 A. koschevnikovi honey bee collection from South Kalimantan. * = samples used for sequencing and analysing in this study (Raffiudin R 24 Januari 2011, personal communication)
No. Sample code Location Southern
latitude
Longitude east
1. AK 1 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.49’ 115 o.17’
2. AK 2 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.49’ 115 o.17’
3. AK 3 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.49’ 115 o.17’
4. AK 4 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.49’ 115 o.17’
5. AK 5 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.50’ 115 o.18’
6. AK 6 HSS Kec. Losado, Kab. Hulu Sungai Selatan 02o.49’ 115 o.24’
7. AK 7 HSS Kec. Losado, Kab. Hulu Sungai Selatan 02o.49’ 115 o.25’
8. AK 8 HSS* Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.49’ 115 o.17’
9. AK 1 BL* Kec. Lanmpihong, Kab. Balangan 02o.20’ 115 o.25’
10. AK 2 BL Kec. Lanmpihong, Kab. Balangan 02o.20’ 115 o.25’
11. AK 3 BL Kec. Lanmpihong, Kab. Balangan 02o.20’ 115 o.25’
12. AK 4 BL Kec. Lanmpihong, Kab. Balangan 02o.20’ 115 o.25’
13. AK 5 BL* Kec. Paringin, Kab. Balangan 02o.20’ 115 o.25’
14. AK 9 HSS Kec. Padang Batung, Kab. Hulu Sungai Selatan 02o.47’ 115 o.17’
15. AK 10 HSS Kec. Losado, Kab. Hulu Sungai Selatan 02o.49’ 115 o.25’
16. AK 11 HSS Kec. Losado, Kab. Hulu Sungai Selatan 02o.49’ 115 o.25’
17. AK 1 HST* Kec. Batu Benawa, Kab. Hulu Sungai Tengah 02o.38’ 115 o.26’
18. AK 2 HST Kec. Batu Benawa, Kab. Hulu Sungai Tengah 02o.38’ 115 o.25’
19. AK 3 HST Kec. Batu Benawa, Kab. Hulu Sungai Tengah 02o.38’ 115 o.25’
20. AK 4 HST Kec. Alai Utara, Kab. Hulu Sungai Tengah 02o.31’ 115 o.27’
21. AK 1 TB* Kec. Batu Licin, Kab. Tanah Bumbu 03º.29’ 116º.00’
22. AK 2 TB Kec. Batu Licin, Kab. Tanah Bumbu 03º.29’ 115º.59’
23 AK 1 KB Kec. Sungai Bali, Kab. Kota Baru 03º.30’ 116º.21’
24 AK 2 KB* Kec. Sungai Bali, Kab. Kota Baru 03º.29’ 116º.21’
25 AK 3 KB Kec. Sungai Bali, Kab. Kota Baru 03º.14’ 116º.15’
26 AK 4 KB Kec. Berangas, Kab. Kota Baru 03º.14’ 116º.13’
27 AK 5 KB Kec. Berangas, Kab. Kota Baru 03º.14’ 116º.13’
28 AK 6 KB Kec. Berangas, Kab. Kota Baru 03º.14’ 116º.13’
Appendix 2 Chromatogram of A. koschevnikovi cytochrome oxidase subunit 1 (COI) gene from Ak8 HSS sample using AmF_cox1 forward primer
BioEdit v ersion 7.0.9.0 (6/27/07)
GT TGATACA 750
CGAGCT TAT T
760
T TACT TCTGC
770
A ACTATA ATC
780
AT TGCTGTAC
790
CA ACAG GA A A
Reverse primer Edited manually
BIO TRACE
BioEdit v ersion 7.0.9.0 (6/27/07) Model 3730 KB.bcp
6258000-04 6258002-04 6258003-03 6258005-00 File: 1st_BASE_262661_1_F.ab1
262661_1_F Lane 10
Signal G:345 A:576 T:654 C:369 KB_3730_POP7_BDTv3.mob ?? no 'MTXF' field
Points 2271 to 15634
Page 1 of 5 4/9/2010
Spacing: 13.0124282836914
G GCATACGT
10
CAT TC T T TC T
20
C CAAGCACAT
30
GCAT T TCCTA 40
TAAT T T T T T T
50
TATAGT TATA 60
C CAT T T T TA A 70
T TG GAG GAT T
80
TG GTA AT TGA
90
T TA AT TC CAT
100
TA ATACT TG G 110
ATCTC CAGAT
120
ATAGCT T T TC
130
CTCGTATA A A 140
TA ACAT TAGA 150
T T T TGAT TAT
160
TAC CAC CT TC
170
AT TAT TA ATA 180
Appendix 3 Chromatogram of A. koschevnikovi cytochrome oxidase subunit 1 (COI) gene from Ak 8HSS sample using AmR_cox1 reverse primer
BIO
TRACE
BioEdit v ersion 6.0.7 (5/19/04)Model 3730 KB.bcp
6258000-04 6258002-04 6258003-03 6258005-00 File: 1st_BASE_262662_1_R.ab1
262662_1_R Lane 8
Signal G:343 A:654 T:641 C:31 KB_3730_POP7_BDTv3.mob ?? no 'MTXF' field Points 2287 to 15458
C C C CAG GAT
10
CATGA AT TAG 20
CA ATGATCA A
30
ATCTATA ATA
40
CTAT TGTCAC
50
A AGACATGCA
60
T T T T TA ATA A
70
T T T T T T T TAT
80
AGT TAT
ACT TG GATC
140
TC CAGATATA
150 GCT T T TC CTC
160 GTATA A ATA A
170
CAT TAGAT T T
180
TGAT TAT TAC
190
CAC CT TCAT T
200
AT TA ATAT TA
210
T TAT TA AGA A
220
AT T TAT T T TA
230
C C CA AGAC CA
24 G
T T T TAC C
620
CG GAT T TG GA
630
T TA AT T TCTC
640
ATAT TGTA AT
650
A A ATGA A AGA
660 GGA A A A A A AG
670
A A AT T T TTG G 680
A A AT T TAGGA
690
ATA AT T TATG 700
CA ATAT TAGG 710
A AT TGGAT TT
720
CTAG GAT T
DZULFAQOR
DEPARTMENT OF BIOLOGY
FACULTY OF MATHEMATICS AND NATURAL SCIENCE
BOGOR AGRICULTURAL UNIVERSITY
Apis koschevnikovi in South Kalimantan. Supervised by RIKA RAFFIUDIN and MOCHAMAD CHANDRA W.
Deforestation in Kalimantan forest by logging and conversion to palm oil plantation destroyed many living organism and carbon-rich ecosystems. This condition was a serious threat for native honeybee species from Kalimantan i.e. Apis koschevnikovi (Hymenoptera; Apidae). Recent observations shows that the population of A. koschevnikovi has decreased persistently. Ecological integrity of A. koschevnikovi database need to be preserved, including molecular database. Cytochrome oxidase subunit I (COI) in mitochondrial DNA (mtDNA) can assist by providing suitable molecular marker with DNA barcode-based technology. Currently, there are 15 haplotypes of COI gene of A. koschevnikovi has published in GeneBank database. The objective of this research was to explore the nucleotide variations of A. koschevnikovi in South Kalimantan based on COI gene. Five regencies were observed in South Kalimantan forest; Hulu Sungai Selatan (HSS), Hulu Sungai Tengah (HST), Kotabaru (KB), Tanah Bumbu (TB), and Balangan (BL). Based on alignment of 738 bp of COI A. koschevnikovi, there were 5 haplotypes of nucleotides were found. The common haplotypes found in TB and HST samples, while others specific in other places. None of the samples showed the same sequence with the nucleotide sequences of GeneBank database, hence these five haplotypes were concur to be new haplotypes.
Keywords : DNA barcode, COI, A. koschevnikovi, mitochondrial DNA, haplotype.
ABSTRAK
DZULFAQOR. Variasi Gen Sitokrom Oksidase subunit I (COI) di DNA Mitokondria pada Apis koschevnikovi di Kalimantan Selatan. Dibimbing oleh RIKA RAFFIUDIN dan MOCHAMAD CHANDRA W.
Deforestasi di hutan Kalimantan yang berupa pembalakan dan konversi hutan menjadi perkebunan kelapa sawit menghancurkan habitat organisme dan ekosistem. Kondisi ini merupakan ancaman yang serius terhadap spesies asli yang berasal dari Kalimantan, termasuk Apis koschevnikovi (Hymenoptera; Apidae). Penelitian-penelitian sebelumnya menunjukkan kecendrungan populasi A. koschevnikovi yang terus menurun. Memelihara integritas ekologi A. koschevnikovi sangat diperlukan, termasuk database molekulernya. Sitokrom Oksidase subunit I (COI) di DNA mitokondria (DNAmt) dapat membantu dengan menyediakan penanda molekuler dengan teknologi berbasis DNA barcoding. Saat ini, telah ditemukan sebanyak 15 jenis haplotipe gen COI dari A. koschevnikovi yang telah dipublikasikan di Genbank. Tujuan dari penelitian ini adalah untuk mengetahui variasi genetik dari A. koschevnikovi Kalimantan Selatan berdasarkan gen COI dengan menggunakan metode sekuensing DNA. Contoh yang digunakan dalam penelitian ini berasal dari 5 kabupaten di Kalimantan Selatan; Hulu Sungai Selatan (HSS), Hulu Sungai Tengah (HST), Kotabaru (KB), Tanah Bumbu (TB), and Balangan (BL). Pengurutan nukleotida dari contoh yang digunakan menghasilkan panjang nukleotida sebanyak 738 bp, dan sebanyak 5 haplotipe dari nukleotida telah ditemukan. Haplotipe umum ditemukan dari contoh yang berasal dari Kabupaten HST dan TB. Hasil pengurutan dari contoh ini tidak menunjukkan kesamaan dengan 15 haplotipe dari Genebank, sehingga 5 haplotipe yang ditemukan dalam penelitian ini dapat disimpulkan sebagai haplotipe baru.
INTRODUCTION
Background
Deforestation in Kalimantan due to logging and forest conversion to palm oil plantation destroyed many living organism and carbon-rich ecosystems (Curran et al.
2004). This condition was a serious threat for the species that is native in Kalimantan such as Apis koschevnikovi,a cavity nesting honey bee. It is classified in the order of Hymenoptera, suborder of Apocrita, superfamily of Apoidea, and family of Apidae (Michener 2000). The distribution of A. koschevnikovi has establishes many new site localities but all within the previously confirmed limits of the tropical evergreen forest of Sundaland (Hadisoesilo et al. 2008). Nowadays, recent observations shows that the population of A. koschevnikovi has decreased persistently due to forest loss and establishment of some clear land to make oil palm, rubber, and coconut plantations (Otis 1991).
Preserving the ecological integrity of A. koschevnikovi database is necessary, including molecular database. Cytochrome oxidase subunit I (COI) in mitochondrial DNA (mtDNA) provide suitable molecular marker with DNA barcode-based technology due to their characteristic i.e no deletion and insertion in its sequence, and slightly variances in their sequences (Hebert et al.
2003). In animals mtDNA, COI usually composed by over 600 bp of long, and at least 648 bp of long is required to be submitted into Genebank database (www.ncbi.nlm.nih.gov).
Currently, there are 15 haplotypes of COI gene of A. koschevnikovi has published in Genebank under Accesion number AF153110 - AF153111, AY012723, AY754729 - AY754732, and DQ016097 - DQ016104 (www.ncbi.nlm.nih.gov). Most of the haplotypes occured from Sabah (haplotype 3-7, hap 11-15), the others are from Sarawak (Malaysia), and West Kalimantan. However, out of 15 haplotypes, three found in Kalimantan. This study was aimed to explore further to find another variations of COI haploytpes for building the database especially in Kalimantan forest.
Research Objective
The objective of this research was to explore the variations of COI gene in
mitochondrial DNA of A. koschevnikovi in South Kalimantan.
MATERIALS AND METHODS
Time and Place
The research was conducted on January 2010 up to February 2011, and it was taken place in Division of Animal Function and Behaviour, Department of Biology, Bogor Agricultural University (IPB).
Honey Bee Collection and DNA Extraction
The objects of my research were DNA from several A. koschevnikovi honey bees. Honey bees used in this study were collection of Dr. Rika Raffiudin (Dept. Biology IPB) that were collected from Kabupaten Hulu Sungai Selatan (Ak HSS), Hulu Sungai Tengah (Ak HST), Balangan (Ak BL), Tanah Bumbu (Ak TB), and Kotabaru (Ak KB) in South Kalimantan (Figure 1). Several worker honey bee from several colonies were collected from each regencies and were preserved in absolute ethanol.
DNA extraction has carried out by using 0.2% Cetyl Trimethyl Ammonium Bromide (CTAB) and ethanol precipitation (Sambrook
et al. 1989).
COI Gene Amplification
INTRODUCTION
Background
Deforestation in Kalimantan due to logging and forest conversion to palm oil plantation destroyed many living organism and carbon-rich ecosystems (Curran et al.
2004). This condition was a serious threat for the species that is native in Kalimantan such as Apis koschevnikovi,a cavity nesting honey bee. It is classified in the order of Hymenoptera, suborder of Apocrita, superfamily of Apoidea, and family of Apidae (Michener 2000). The distribution of A. koschevnikovi has establishes many new site localities but all within the previously confirmed limits of the tropical evergreen forest of Sundaland (Hadisoesilo et al. 2008). Nowadays, recent observations shows that the population of A. koschevnikovi has decreased persistently due to forest loss and establishment of some clear land to make oil palm, rubber, and coconut plantations (Otis 1991).
Preserving the ecological integrity of A. koschevnikovi database is necessary, including molecular database. Cytochrome oxidase subunit I (COI) in mitochondrial DNA (mtDNA) provide suitable molecular marker with DNA barcode-based technology due to their characteristic i.e no deletion and insertion in its sequence, and slightly variances in their sequences (Hebert et al.
2003). In animals mtDNA, COI usually composed by over 600 bp of long, and at least 648 bp of long is required to be submitted into Genebank database (www.ncbi.nlm.nih.gov).
Currently, there are 15 haplotypes of COI gene of A. koschevnikovi has published in Genebank under Accesion number AF153110 - AF153111, AY012723, AY754729 - AY754732, and DQ016097 - DQ016104 (www.ncbi.nlm.nih.gov). Most of the haplotypes occured from Sabah (haplotype 3-7, hap 11-15), the others are from Sarawak (Malaysia), and West Kalimantan. However, out of 15 haplotypes, three found in Kalimantan. This study was aimed to explore further to find another variations of COI haploytpes for building the database especially in Kalimantan forest.
Research Objective
The objective of this research was to explore the variations of COI gene in
mitochondrial DNA of A. koschevnikovi in South Kalimantan.
MATERIALS AND METHODS
Time and Place
The research was conducted on January 2010 up to February 2011, and it was taken place in Division of Animal Function and Behaviour, Department of Biology, Bogor Agricultural University (IPB).
Honey Bee Collection and DNA Extraction
The objects of my research were DNA from several A. koschevnikovi honey bees. Honey bees used in this study were collection of Dr. Rika Raffiudin (Dept. Biology IPB) that were collected from Kabupaten Hulu Sungai Selatan (Ak HSS), Hulu Sungai Tengah (Ak HST), Balangan (Ak BL), Tanah Bumbu (Ak TB), and Kotabaru (Ak KB) in South Kalimantan (Figure 1). Several worker honey bee from several colonies were collected from each regencies and were preserved in absolute ethanol.
DNA extraction has carried out by using 0.2% Cetyl Trimethyl Ammonium Bromide (CTAB) and ethanol precipitation (Sambrook
et al. 1989).
COI Gene Amplification
P. Sebuku minutes (mins) at 940 C for initial denaturing, 30
Table 1 Nucleotide primers for amplifying COI gene of A. koschevnikovi and primer positions based on mitochondrial DNA of Apis mellifera (Crozier & Crozier 1993)
minutes (mins) at 940 C for initial denaturing, 30 cycles of 1 min at 940 C (denaturing), 1 min and 30 seconds at 550 C (annealing), 2 mins at 720 C (DNA elongation) and followed by 5 mins for final extension.
Electrophoresis and DNA Visualization
Amplified DNA was migrated using 6% Polyacrilamid Gel Electrophoresis (PAGE) which was submerged in 1x TBE buffer (10 mM Tris-HCl, 1 M Borat acid, and 0.1 mM EDTA), operated for 50 minutes at 200 V. The compositions of 6% gel acrilamide were 12 ml of aquades, 4 ml of 5xTBE, 4 ml of acrilamide, 15 µl of TEMED, and 160 µl of 10% APS.
DNA visualization used sensitive Silver Nitrate staining (Tegelstrom 1986). Gel resulting from electrophoresis was washed using CTAB solution (0.2 g/200 ml of aquades) for 8 mins, and was flushed using 200 ml of aquades for 2 mins (2 times). Subsequently, its gel was submerged in NH4OH solution (2.4 ml/200 ml of aquades)
for 8 mins, and solution of mixture contained 0.32 g of AgNO3, 0.8 ml of NH4OH, 80 µl
NaOH, and 200 ml of aquades for 10 mins. Then, the gel was flushed again using the same condition as above, and was submerged again in the solution of mixture contained 4 g of NA2CO3, 100 µl of formaldehida, and 200
ml of aquades until the bands of DNA seen. Staining process was stopped using 200 µl of 1% acetat acid in 200 ml of aquades.
Sequencing and DNA Analysis
Genetic variations in COI mitochondrial region was analysed based on DNA sequencing method. DNA was sequenced by using ABI 3100 and 3730XL sequencer machine at sequencing company. Nucleotide sequence were edited and analysed using Genetyx Win Version 4.0 software. Forward and reverse sequence were aligned to obtain contiguous sequence by using Clustal X software (Thompson et al. 1997). All DNA contiguous sequences was aligned together with haplotypes databases of COI gene of A. koschevnikovi in Genebank NCBI (National Center for Biotechnology Information) under Accesion number AF153110-AF153111, Primer Nucleotide sequence of primer
(5’-3’) (Crozier & Crozier 1993)
Positions primer in mitochondrial DNA of Apis mellifera
(Crozier & Crozier 1993) Forward amF_cox1 CCCCAGGATCATGAATTAGC 1915-1934 Reverse amR_cox1 TCATTGCTGTACCAACAGGAA 2719-2739 Figure 1 Sampling location for A. koschevnikovi COI gene in South Kalimantan as shown
by black square (Raffiudin R 24 Januari 2011, personal communication). Number = colonies number (refer to Appendix 1).
P. Laut
AY012723, AY754729- AY754732, and DQ016097-DQ016104
(www.ncbi.nlm.nih.gov) (Table 2).
Edited DNA sequences were translated into putative amino acid using Genetyx and were aligned by using Clustal X software.
DNA genetic distances and phylogenetic analysis were constructed between current samples and 15 haplotypes in Genebank as ingroup while A. cerana with accesion number GQ162109, and A. mellifera with accesion number L06178 as outgroups. These analysis was carried out by using MEGA4 software based on Kimura-2-parameter substitution method and Neighbour Joining
(NJ) construction with 1000x bootstrap (Tamura et al. 2007).
RESULT
DNA Amplification
There were 29 samples of A. koschevnikovi
COI gene from five regencies in South Kalimantan were amplified using AmF_cox1 and AmR_cox1 primer (Table 3). All samples showed the same results i.esingle band with approximately 900 bp of nucleotide (Figure 2).
Table 2 A. koschevnikovi COI gene in published Genebank database (www.ncbi.nlm.nih.gov)
No Accesion
number Author Haplotypes Source
Nucleotides long (bp)
1 AF153110 Tanaka, H. Roubik, Kato,
M., Liew, F., and Gunsalam, haplotype 1 Brunei 1041
2 AF153111
Tanaka, H. Roubik, Kato, M., Liew, F., and Gunsalam, G.
haplotype 2 Malaysia: Lamibir,
Sarawak 1041
3 AY012723 Tanaka, H. , Suka T.,
Roubik, and Mohamed M. haplotype 3
Malaysia: Crocker
Range Park, Sabah 1041
4 AY754729 Suka, T., and Tanaka, H. haplotipe 4 Malaysia: Crocker
Range Park, Sabah 1041
5 AY754730 Suka, T., and Tanaka, H. haplotipe 5 Malaysia: Crocker
Range Park, Sabah 1041
6 AY754731 Suka, T., and Tanaka, H. haplotipe 6 Malaysia: Crocker
Range Park, Sabah 1041
7 AY754732 Suka, T., and Tanaka, H. haplotipe 7 Malaysia: Crocker
Range Park, Sabah 1041
8 DQ016097
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 8
Indonesia: kutai national park, east Kalimantan
1041
9 DQ016098
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 9 Indonesia: Loksad,
south Kalimantan 1041
10 DQ016099
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 10
Indonesia: field site near from sadap, west Kalimantan
1041
11 DQ016100
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 11 Malaysia: Tawau,
Sabah 1041
12 DQ016101
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 12 Malaysia: Tawau,
Sabah 1041
13 DQ016102
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotipe 13 Malaysia: Tawau,
Sabah 1041
14 DQ016103
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotype 14 Malaysia: Tawau,
Sabah 1041
15 DQ016104
Tanaka, H. , Suka, T., Kahono, S., Samejima, H. , Maryati, M., and Roubik.
haplotype 15 Malaysia: Tawau,
AY012723, AY754729- AY754732, and DQ016097-DQ016104
(www.ncbi.nlm.nih.gov) (Table 2).
Edited DNA sequences were translated into putative amino acid using Genetyx and were aligned by using Clustal X software.
DNA genetic distances and phylogenetic analysis were constructed between current samples and 15 haplotypes in Genebank as ingroup while A. cerana with accesion number GQ162109, and A. mellifera with accesion number L06178 as outgroups. These analysis was carried out by using MEGA4 software based on Kimura-2-parameter substitution method and Neighbour Joining
(NJ) construction with 1000x bootstrap (Tamura et al. 20