MOLECULAR IDENTIFICATION OF
Fusarium
spp.
ASSOCIATED WITH EAGLEWOOD BASED ON ITS
INTERNAL TRANSCRIBED SPACER SEQUENCE
RURI PRIHATINI ARIMBI
DEPARTMENT OF BIOLOGY
FACULTY OF MATHEMATICS AND NATURAL SCIENCE BOGOR AGRICULTURAL UNIVERSITY
STATEMENT OF MINI
-
THESIS AND SOURCE OF
INFORMATION AND DEVOLVING COPYRIGHT *
I hereby declare that mini-thesis entitled Molecular Identification of
Fusarium spp. Associated with Eaglewood Based on Its Internal Transcribed Spacer Sequence is a true work of me with the direction of the supervisor and has not been submitted in the form of anything to any college. A source of information derived or quoted of works issued or published or not published from other authors mentioned in the text and listed in the bibliography at the end of this dissertation.
I hereby assign copyright of my paper to the Bogor Agricultural University.
Bogor, January 2014
Ruri Prihatini Arimbi
2
ABSTRACT
RURI PRIHATINI ARIMBI. Molecular Identification of Fusarium spp. Associated with Eaglewood Based on Its Internal Transcribed Spacer Sequence. Under supervision GAYUH RAHAYU and UTUT WIDYASTUTI.
Eaglewood was thought to be formed through fungal infection. Acremonium
and Fusarium were able to induce symptom of eaglewood formation in 2 year-old eaglewood trees (Aquilaria crassna, A. malaccensis, and A. microcarpa). The objective of this research is to identify nine fungal isolates from eaglewood that morphological character tentatively determined belonging to Acremonium and
Fusarium from eaglewood by molecular approach using its Internal Transcribed Spacer region. The primers ITS 1 and ITS 4 had successfully amplified the ITS region (500-700 bp) of all investigated strains. Fungal isolates from eaglewood comprises of 1 isolate Fusariumacaciaemearnsii, 1 isolate Fusariumoxysporum, 6 isolate Fusarium solani complex and 1 isolate Fusarium solani fsp. Batatas. The molecular identification was not supported the morphological identification. Based on molecular analysis, the Acremonium-like morphological genus was apparently Fusarium sp. in the Section Martiella sensu Booth 1971.
Keywords: eaglewood clump, Fusarium, ITS primer.
ABSTRAK
RURI PRIHATINI ARIMBI. Molecular Identification of Fusarium spp. Associated with Eaglewood Based on Its Internal Transcribed Spacer Sequence. Dibimbing oleh GAYUH RAHAYU dan UTUT WIDYASTUTI.
Gaharu dapat terbentuk melalui infeksi cendawan. Acremonium dan
Fusarium mampu menyebabkan gejala pembentukan kayu gaharu di pohon-pohon gaharu berusia 2 tahun (Aquilaria crassna, A. malaccensis, dan A. microcarpa). Tujuan dari penelitian ini ialah mengidentifikasi 9 isolat cendawan asal pohon gaharu yang secara morfologi termasuk Acremonium dan Fusarium melalui pendekatan molekuler berdasarkan daerah Internal Transcribe Spacer-nya. Daerah ITS (500-700 bp) semua isolat telah berhasil diamplifikasi oleh primer ITS 1 dan ITS 4. Cendawan yang berasal dari gaharu terdiri dari 1 isolat Fusarium acaciaemearnsii, 1 isolat Fusariumoxysporum, 6 isolat Fusariumsolani complex dan 1 isolat Fusarium solani fsp. Batatas. Identifikasi molekuler tidak mendukung identifikasi sementara cendawan secara morfologi. Berdasarkan analisis molekuler cendawan yang secara morfologi mirip Acremonium adalah
3
An Undergraduate Thesis Intended to Acquire Bachelor Degree
In
Departement of Biologi
MOLECULAR IDENTIFICATION OF
Fusarium
spp.
ASSOCIATED WITH EAGLEWOOD BASED ON ITS
INTERNAL TRANSCRIBED SPACER SEQUENCE
RURI PRIHATINI ARIMBI
DEPARTMENT OF BIOLOGY
FACULTY OF MATHEMATICS AND NATURAL SCIENCES BOGOR AGRICULTURAL UNIVERSITY
5
Title : Molecular Identification of Fusarium spp. Associated with Eaglewood Based on Its Internal Transcribed Spacer Sequence Name : Ruri Prihatini Arimbi
NIM : G34060327
Approved by
Dr Ir Gayuh Rahayu Supervisor I
Dr Ir Utut Widyastuti, MSi Supervisor II
Endorsed by
Dr Ir Iman Rusmana, M. Si Head of Department
7
PREFACE
This mini-thesis is made through a research entitled Molecular Identification of Fusarium spp. Associated with Eaglewood Based on Its Internal Transcribed Spacer Sequence. It is intended to confirm the identification that was made by morphological approach. Hopefully, it might be useful information for readers and science development especially in eaglewood project.
I would firstly thank to Dr. Gayuh Rahayu and Dr. Ir. Utut Widyastuti, MSi. as my supervisors. They gave me a lot of trust and flexibility on the project so I could dealt with such a challenging project, I could collect data in a short time and above all complete this project. I am thanking them for the great support.
I would also like to thank to all staffs and lecturers in RCBio and IPBCC for all knowledge and time they shared to me. To all Biology 43, thank you for all the best time we spent together. At last, I would like to sent acknowledgement to Bu Sri Listyowati and to Uncle Jo for helping me personally in facing all the problems while making this mini-thesis.
Bogor, November 2013
8
Data Analysis Procedures ... 11
Maintenance of working culture ... 11
DNA extraction and PCR amplification ... 11
Electrophoresis ... 12
DNA sequencing and Analysis ... 12
Time and Place ... 12 1 Microscopic features of conidial heads of Acremonium IPBCC 07.563 ... 13
2 The position of Internal Transcribed Spacer gene segment ... 13
3 Electrophoresis DNA fragment of nine fungal isolates from eaglewood ... 13
3 Molecular phylogenetic tree ... 15
LIST OF TABLE Page 1 Strain isolates from eaglewood and the references were used in this study ... 10
9
INTRODUCTION
Background
Eaglewood is a non-timber forest product with high economy value especially from its fragrant resin deposit. This resinuous substance is accumulated in wood tissue as a response toward wounding or pathogens infection. According to Nobuchi and Siripatanadilok (1991), eaglewood clump was thought to be formed through fungal infection. Various species of Fusarium such as Fusarium oxysporum, Fusarium bulbigenium, and Fusarium lateritium have been isolated by Santoso (1996). Rahayu et al. (1999) stated that several isolates of
Acremonium sp. From eaglewood clump of Gyrinops versteegii and Aquilaria malaccensis were able to induce symptom of clumps formation in 2 year-old eaglewood trees (Aquilaria crassna, Aquilaria malaccensis, and Aquilaria microcarpa). IPB Culture Collection (IPBCC) has a number of collection of mitosporic fungi from eaglewood that have been identified tentatively on morphological bases to either Acremonium or Fusarium (Gayuh Rahayu, personal communication 14 February 2013).
Acremonium and Fusarium are closely related genera which are differentiated on the bases of the presence of macroconidia. Acremonium lacks macroconidia and Fusarium have macroconidia. The genus Acremonium
(Summerbell et al. 2011) includes some of the most simply structured of all filamentous anamorphic fungi. The characteristic morphology consists of septate hyphae giving rise to thin, tapered, mostly lateral phialides produced singly or in small groups conidia. Conidia tend to be unicellular, produced in mucoid heads or unconnected chains. They can be hyaline or melanised, but the hyphae are usually hyaline. Colonies can produce conidia in less than a week in a warm, moist environment, making Acremonium a very fast growing fungus. The colony typically has a slightly powdery texture, and a color which can vary from gray to pink.
Fusarium colonies are also usually fast growing, pale or brightly colored (depending on the species) and may or may not have a cottony aerial mycelium. The color of the mycelia varies from whitish to yellow, brownish, pink, reddish or lilac shades. These fungi is a large genus and widely distributed in soil and in association with plants. Most species are harmless saprobes and relatively abundant members of the soil microbial community, but some are phytopatogenic and human pathogens (Summerbell and Schroers 2002).
10
phylogenetically considered as a variant of Fusarium solani complex based on their 28S rDNA.
Accurate identification of these fungi is, therefore, essential prior to broad application of these fungi for artificial inoculation and provide IPBCC with information for building up a molecular reference for fungal identification.
Objective conventional methods or morphological character. Accurate identification using molecular approach of these fungi is essential to determine appropriate phylogenic and discover its genetic information.
METHODS
Materials
Materials used in this research were 9 fungal isolates from eaglewood (Table 1) and 1 isolate of Fusarium oxysporum from Cucumis sativus IPBCC 88.012 (CBS 254.52), PDA (Potato Dextrose Agar) media, PDB (Potato Dextrose Broth) media, alcohol (70%), antibiotics cloramfenicol, chemical for DNA extraction, primers ITS 1 (reverse primer) and ITS 4 (forward primers).
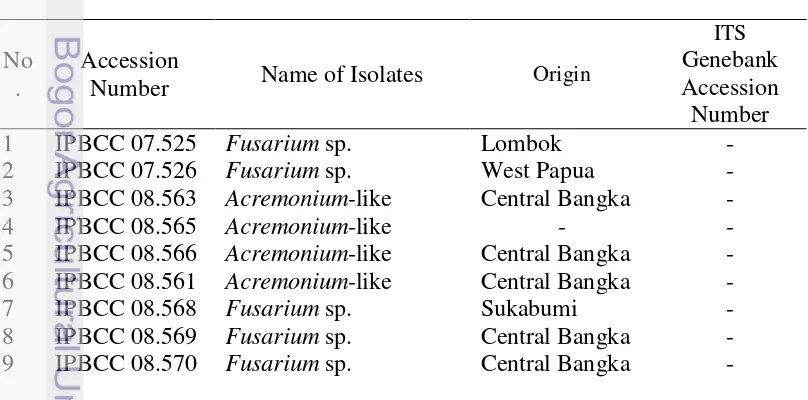
Table 1. Strain isolates from eaglewood and the references were used in this study
No .
Accession
Number Name of Isolates Origin
11
10 IPBCC Fusarium oxysporum Cucumissativus -
11 Isolate By125 F.oxysporum - GQ365156.1
12 NRRL 42499 F.oxysporum - DQ790539.1
13 NRRL 13459 F.concolor - GQ505763.1
14 NRRL 26419 F.equiseti - GQ505688.1
15 - Fusariumpoae - JQ912669.1
16 NRRL34207 F. acaciae mearnsii - DQ459854.1
17 NRRL 52789 F.acuminatum - JF740933.1
18 ISPaVe 2010 F.lateritium - FN547465.1
19 NRRL 34033 F. brachygibbosum - GQ505450.1
20 NRRL 28505 F.nelsonii - GQ505436.1
21 NRRL 34014 Fusarium sp. - GQ505441.1
22 CBS 116522 F.delphinoides - EU926242.1
23 CBS 116521 F.dimerum - EU926259.1
33 P266_D1_10 Acremonium furcatum - JF311914.1
34 Isolate 1125 A. atrogriseum - JX847767.1
Equipment
Equipments used in this research i.e. a compound microscope, bio-safety cabinet, autoclave, micropipettes, electrophoresis and PCR machine.
Data Analysis Procedures
Maintenance of Working Culture
Culture stock of Acremonium sp. and Fusarium sp. were subcultured onto PDA and incubated at room temperature for five to seven days. These cultures were then used to obtain single spore cultures. A small piece of mycelium originating from a single spore was then cut off and transferred to new PDA plates to obtain working cultures. From each working cultures, three pieces of mycelia plug were transferred on to PDB and incubated for 3 days in shaker incubator. Actively growing mycelia were used as a source of DNA genom.
DNA Extraction and PCR Amplification
DNA genome was extracted from mycelia using CTAB method of Sambrook et al. (1989) with modification in PVP (Polivinil Pirolidon) 1%. Genomic DNA were amplified using MJ Research PCR thermal cycle machine
12
mix 2mM 1 µL, TE buffer 1 µL, MgCl 0.2 µL, and Taq DNA Polymerase enzyme 0.1 µL, 0.25 µL primers and 0.5 µL DNA template. Amplification PCR was performed for 35 cycles with an initial denaturation period of 3 minutes at 94oC, denaturation period of 30 seconds at 94oC, annealing for 30 seconds, at 55oC, charged molecules which simulates DNA fragments of a certain size and length. It was placed in the well of an electrophoresis gel. Electrophoresis was run at 70 volt for 45 minutes. In order to confirm insert size based on DNA marker, the PCR Analysis) V. Phylogenetic construction were made included the reference strain of
Fusarium sp. from the genebank (Table 1) using Maximum likelihood in MEGA V. Acremoniumatrogriseum, Acremonium furcatum and Trichoderma harzianum
were used as an outgroup taxa. The robustness of the phylogenetic tree was estimate bootstrap analysis with 1.000 replications. The tree was drawn in Adobe Reader.
Time and Place
This research was conducted from May 2010 until July 2011 in Research Center for Bioresources and Biotechnology (RCBio), Bogor Agricultural University.
RESULT AND DISCUSSION
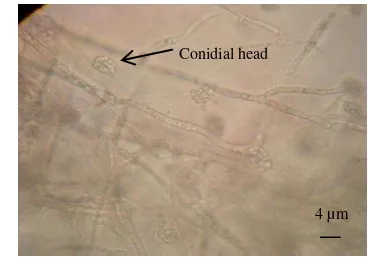
Isolate IPBCC 08.568 and IPBCC 08.570 were typically has Fusarium
characteristics in which the macroconidia and microconidia are present, with a few macroconidia. Whilst the microscopic characters of the other isolates are typically Acremonium like (Figure 1). They have no macroconidia. The microconidia of these isolates are produced in a slimy head of a simple phialide. This trait closed to Fusarium solani in the Section Martiella of Booth (1971). Such confusion was also discussed by Summerbell and Schroers (2002) on
13
Fig 1. Microscopic features of conidial heads of Acremonium IPBCC 08.563 The molecular identification was not supported the morphological identification. The primers had successfully amplified the ITS region about 500-700 bp (Figure 2) of all investigated strains and the DNA fragment was visualized using UV transluminator (Figure 3). All specimens either with or without macroconidia have been identified as Fusarium. A similar finding of Summerbell and Schroers (2002) was Acremonium falciforme is phylogenetically considered as a variant of Fusariumsolanicomplex based on their ITS sequences.
Fig 2. The position of Internal Transcribed Spacer gene segment (Lilley and Chinabut 2000)
9 8 7 6 5 4 3 2 1 Marker (3530 bp) (600bp)
Fig 3. Electrophoresis DNA fragment of nine fungal isolates from eaglewood
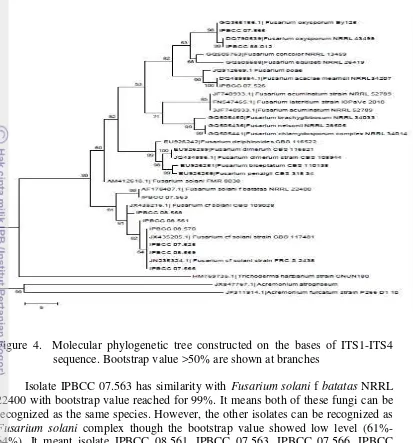
Tree topology (Figure 4) indicated that both Trichoderma and Acremonium are good outgroup, since they formed separate clade with Fusarium. Within
Fusarium there are two clades i.e. mixture species of Fusarium as clade one with 60% bootstrap value and Fusarium solani complex as a clade two with 61%.
Conidial head
14
Clade one can be divided into several subclades. Amongst those is Fusarium oxysporum complex.
In the clade one, Fusarium solani FMR 8C38 seemed to be misplaced, but actually may not. One strain (IPBCC 07.526) that is Fusarium acaciae mearnsii with 100% bootsrap value. These situation indicated that ITS is not a good region to differentiate species of Fusarium. For this purpose, systematicians use Tef-1α for molecular classification of Fusarium such as FusariumID (Geiser et al. 2004, Hsua et al. 2011). mearnsii, Fusarium oxysporum, Fusarium solani complex and Fusarium solani fsp. batatas.
Table 2. Fungal isolates from eaglewood identification result based on ITS region No Accession
Sequence analysis of ITS gene showed two isolates namely IPBCC 07.565 and IPBCC 88.012 has the closest similarity of Fusarium oxysporum complex.
Fusarium oxysporum comprises a group of soil inhabitants that can exist as saprophytes in the soil debris but also as pervasive plant endophytes colonizing the plant roots. It stands in the ground as clamidiospora that can be found on the roots of those who are ill Many strains of these species are pathogenic to plant crops. The Fusarium oxysporum species complex causes catastrophic crop losses in over 150 plant species and is considered one of the world’s most economically significant soil borne plant pathogens. This species is, however, primarily non-pathogenic and forms endophytic, saprophytic, and latent non-pathogenic relationships with its plant hosts (The Royal Botanical Garden 2013).
15
Figure 4. Molecular phylogenetic tree constructed on the bases of ITS1-ITS4 sequence. Bootstrap value >50% are shown at branches
Isolate IPBCC 07.563 has similarity with Fusariumsolani f batatas NRRL 22400 with bootstrap value reached for 99%. It means both of these fungi can be recognized as the same species. However, the other isolates can be recognized as
Fusarium solani complex though the bootstrap value showed low level (61%-64%). It meant isolate IPBCC 08.561. IPBCC 07.563. IPBCC 07.566, IPBCC 07.568, IPBCC 08.569 and IPBCC 08.570 nearly close to Fusariumsolani species than any other Fusarium species. Fusarium solani is one of the most frequently isolated fungi from soil and plant debris ubiquitous in soil and also decaying plant material, where they act as decomposers. However, they are also host-specific pathogens of a number of agriculturally important plants, including pea, cucurbits, and sweet potato (Zhang et al, 2006).
Acremonium were used as an outgroup for these isolates. According to Thomas et al. (2003), the similarities between Fusarium and Acremonium may produce single-celled conidia of similar shapes on erect phialides, which were a characteristic of the case isolate. While Fusarium also produces sickle-shaped multicellular macroconidia in sporodochia, these are not always observed in the laboratory. Colonies of Fusarium sp. often produce various shades of red, blue, or purple, but these can be absent or subtle. Furthermore, Acremonium sp. may produce similar pigments (Thomas et al. 2003).
16
fungi that are highly interactive in root, soil, and foliar environment (Gary et al. 2004). They show a high level of genetic diversity, and can be used to produce a wide range of products of commercial and ecological interest. They are prolific producers of extracellular proteins, and are best known for their ability to produce enzymes that degrade cellulose and chitin, although they also produce other useful enzymes (Gary et al. 2004).
CONCLUSION
Molecular identification on the base of Internal Transcribed Spacer indicated that nine fungal isolates from eaglewood comprises of 1 isolate
Fusarium acaciae mearnsii, 1 isolate Fusarium oxysporum, 6 isolates Fusarium solani complex and 1 isolate Fusariumsolani fsp. Batatas.
REFERENCES
Anwar G dan Hartal. 2007. Teknologi peningkatan kualitas kayu gubal gaharu (Aquilaria malaccensis Lamk.) di kawasan pesisir Bengkulu dengan inokulan jamur penginduksi resin. Jurnal Ilmu-Ilmu Pertanian Indonesia 3: 464 – 471. Choi YW, Hyde KD, and Ho WH. 1999. Single spore isolation of fungi. Fungal
Diversity 3: 29-38.
Darmono TW, Ilyas J, dan Santosa DA. 2006. Pengembangan penanda molekuler untuk deteksi Phytophtora palmivora pada tanaman kakao. Menara Perkebunan 74 (2): 87-96.
Hall TA (1999). BioEdit: a user-friendly biologicalsequence alignment editor and analysis programfor Windows 95/98/NT. Nucleic Acids Symposium Series 41:95-98
Hsuan HM, Salleh B, and Zakaria L. 2011. molecular identification of Fusarium
species in Gibberellafujikuroi species complex from rice, sugarcane and maize from Peninsular Malaysia. International Journal of Molecular Sciences 12: 6722-6732.
Kidd KK, Osterhoff D, Erhard L, Stone WH. 1974. The use of genetic relationships among cattle breeds in the formulation of rational breeding policies: an example with South Devon (South Africa) and Gelbvieh (Germany). Animal Blood Groups and Biochemical Genetics 5: 21-28.
17 Gary et al. 2004. Trichoderma species — opportunistic, avirulent plant symbionts.
Nature Reviews Microbiology 2: 43-56.doi:10.1038.
Geiser DM, Jimenez-Gasco MM, Kang S, Makalowska I, Veeraraghavan N, Ward
TJ, Zhang N, Kuldau GA, and O’Donnell K. 2004. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. European Journal of Plant Pathology 110: 473–479.
Mycology Online. 2010. Acremonium sp. http://www.mycology.adelaide.edu.au [ 30 Jan 2010].
Mycology Online. 2010. Fusarium sp. http://www.mycology.adelaide.edu.au [ 30 Jan 2010].
Nei M and Kumar S. 2000. Molecular Evolution And Phylogenetics. New York: Oxford University Press Inc.
Nobuchi T and Siripatanadilok S. 1991. Preliminary observation of Aquilaria crassna wood with the formation of aloeswood. Bulletin Kyoto University Forest 63: 226-235.
Santoso E. 1996. Pembentukan gaharu dengan cara inokulasi. Turjaman M, editor. In: Makalah diskusi hasil penelitian dalam menunjang pemanfaatan hutan yang lestari; 1996 Maret 11-12. Bogor, Indonesia. Bogor (ID). Pusat Litbang Hutan dan Konservasi Alam. page 1-3.
Summerbell RC and HJ Schroers. 2002. Analysis of phylogenetic relationship of
Cylindrocarpon lichenicola and Acremo-nium falciforme to the Fusarium solani species complex and a review of similarities in the spectrum of opportunistic Infections caused by these fungi. J of Clinical Microbiol. 40 (8): 2866–2875. DOI: 10.1128/JCM.40.8. 2866– 2875.2002.
Summerbell RC et al. 2011. Acremonium phylogenetic overview and revision of
Gliomastix, Sarocladium, and Trichothecium. Studies in Mycology 68: 139-162
18
CURRICULUM VITAE
The author was born in Padang on June 17th 1988 as the first child of Rumzi Jani and Tri Koryani. In 2006, the author was graduated from SMA Negeri 5 Kota Jambi and entered Department of Biology, Faculty of Mathematics and Natural Science, Bogor Agricultural University. In 2008, she conducted Field
Study with the title ―Eksplorasi Rhizobakteria Asal Perakaran Legum dari Kawasan Taman Wisata Alam Situ Gunung‖. In 200λ, she did field work with the title ―Pembiakan Apantheles sp. sebagai Biokontrol Penggerek Batang pada
Tanaman Tebu‖ in PT. PG. Rajawali II at Subang city.
As an academia, author assisted practical class of Basic Biology in 2009 until 2010, Molecular Genetics in 2010, and Mycology in 2010. She became finalist in PKM and in 2010 she also won the competition of student debate in Faculty of Mathematic and Natural Science.