Accepted Manuscript
Title: Ultraviolet B inhibition of DNMT1 activityviaAhR activation dependent SIRT1 suppression in CD4+ T cells from systemic lupus erythematosus patients
Authors: Zhouwei Wu, Xingyu Mei, Zuolin Ying, Yue Sun, Jun Song, Weimin Shi
PII: S0923-1811(17)30002-6
DOI: http://dx.doi.org/doi:10.1016/j.jdermsci.2017.03.006
Reference: DESC 3148
To appear in: Journal of Dermatological Science Received date: 3-1-2017
Revised date: 22-2-2017 Accepted date: 8-3-2017
Please cite this article as: Wu Zhouwei, Mei Xingyu, Ying Zuolin, Sun Yue, Song Jun, Shi Weimin.Ultraviolet B inhibition of DNMT1 activity via AhR activation dependent SIRT1 suppression in CD4+ T cells from systemic lupus erythematosus patients.Journal of Dermatological Sciencehttp://dx.doi.org/10.1016/j.jdermsci.2017.03.006
1
Ultraviolet B inhibition of DNMT1 activity via AhR activation dependent SIRT1 suppression in CD4+ T cells from systemic lupus erythematosus patients
Zhouwei Wu*#, M.D., Ph.D; Xingyu Mei*, M.D.; Zuolin Ying, M.D. Ph.D.; Yue Sun, M.D.;
Jun Song, M.D.; Weimin Shi# M.D. Ph.D.
Department of Dermatology, Shanghai First People’s Hospital, Shanghai Jiaotong University,
Shanghai, China.
* These authors contributed equally to this manuscript.
# Corresponding author: Weimin Shi
Email: shiweimin2@gmail.com
Zhouwei Wu
Email: 418950049@qq.com
Word count: 3171
Number of references: 33
Table: 3
2 Highlights
DNMT1 activity was modulated by SIRT1 expression in CD4+ T cells.
UVB inhibited DNMT1 activity via decreasing SIRT1 expression in lupus CD4+ T
cells.
SIRT1 expression was directly and negatively regulated by AhR.
AhR was involved in the UVB-induced SIRT1/DNMT1 inhibition in lupus CD4+ T
cells.
Abstract
Background: Previous studies have reported that ultraviolet B (UVB) inhibits DNA
methyltransferase1 (DNMT1) activity in CD4+ T cells from systemic lupus erythematosus
(SLE) patients. Silent mating type information regulation 2 homolog 1 (SIRT1) is a type of
Class III histone deacetylases (HDACs), and has been reported to play roles in the
pathogenesis of different autoimmune diseases and can modulate DNMT1 activity. Moreover,
aryl hydrocarbon receptor (AhR) has been reported to link UVB with SLE. However, the
assay, quantitative real-time PCR (qRT-PCR), Western blotting, RNA interference using
small interfering RNA and Chromatin Immunoprecipitation (ChIP) assay were employed.
Results:DNMT1 activity was inhibited in si-SIRT1-transfected CD4+ T cells, and increased
3
of SIRT1 were suppressed by UVB exposure in lupus CD4+ T cells. UVB-inhibited DNMT1
activity was reversed by SRT1720 in control-transfected lupus CD4+ T cells, but not in
si-SIRT1-transfected lupus CD4 + T cells. Furthermore, AhR activation by VAF347 reduced the
mRNA and protein expression of SIRT1. ChIP using an antibody against AhR in normal
CD4+ T cells revealed a 16-fold stronger signal at the site about 1.6 kb upstream from the
translation start site of the SIRT1 promoter. Finally, UVB could activate AhR and inhibit the
mRNA and protein expression of SIRT1. AhR knockdown abrogated the inhibition of
UVB-mediated SIRT1 mRNA and protein expression and DNMT1 activity in lupus CD4+ T cells.
Conclusion: UVB suppressed SIRT1 expression via activating AhR, and subsequently
inhibited DNMT1 activity in CD4+ T cells from SLE patients.
Keywords: systemic lupus erythematosus; aryl hydrocarbon receptor; silent mating type
4 1. Introduction
Systemic lupus erythematosus (SLE) is an autoimmune connective tissue disease
characterized by uncontrolled lymphocyte autoreactivity that triggers inflammation and tissue
damage in many parts of the body. The molecular mechanisms initiating the autoimmune
response are poorly understood, although both genetic and epigenetic factors are
implicated[1].
Studies in humans have shown that DNA hypomethylation is involved in the
pathogenesis of SLE [2]. It is well accepted that DNA methylation is catalyzed by several
types of DNA methyltransferases (DNMTs), and DNMT1 is essential for the maintenance of
global DNA methylation pattern [3]. The abnormal catalytic activity of DNMT1 and
subsequent aberrant DNA methylation status have been identified and shown to play roles in
different human diseases [4, 5]. We also have previously shown that ultraviolet B (UVB)
inhibited DNMT1 catalytic activity in CD4+ T cells in a dosage-dependent manner and
further enhanced the decrease of global DNA methylation in SLE patients [6]. However, the
exact molecular mechanisms by which DNMT1 activity is inhibited by UVB in lupus CD4+
T cells remain largely unknown.
Aryl hydrocarbon receptor (AhR) is a transcription factor that resides in the cytosol of
many different kinds of cells, and is a member of the bHLH-PAS protein family. Activation
of AhR leads to conformational change and translocation to the nucleus where it binds to its
dimerization partner, aryl hydrocarbon receptor nuclear translocator (ARNT). The
AhR/ARNT complex initiates transcription of genes with promoters containing a
dioxin-responsive element (DRE) consensus sequence [7]. Interestingly, studies indicate that AhR
activation plays diverse roles in the regulation of the immune system in different immune
cells, including those in T and B cells, mucosal cells, antigen-presenting cells, including
5
in autoimmune disorders has generated significant interest. It was reported that AhR
activation regulated the methylation status of FoxP3 and IL-17 gene promoters and
ameliorated experimental colitis [10]. Wide range factors can activate AhR including
2,3,7,8-Tetrachlorodibenzodioxin (TCDD), tryptophan derivatives, flavonoids, biphenyls and UV
radiation [11]. Recent study showed that a significant AhR activation was observed in the
peripheral blood of active SLE patients, and UV enhanced this effect by converting
propranolol, a potential lupus-inducing drug into a proinflammatory AhR ligand [12].
However, the relationship between UVB and AhR in lupus CD4+ T cells is still poorly
understood.
DNMT1 is a multi-domain large protein, comprising ~1,620 amino acids in humans. It
consists of an N-terminal regulatory domain, which mediates nuclear localization and
targeting to replication foci and discriminates between unmethylated and hemimethylated
DNA; a C-terminal catalytic domain; and a central region, which contains a cysteine-rich
Zn-binding motif, a polybromo motif, and a series of repeating glycine-lysine dipeptides (the GK
linker) [13]. A number of intrinsic and extrinsic factors participate in the regulation of the
expression, stability and activity of this enzyme [14]. Previous work demonstrated that
sumoylation, phosphorylation, methylation, and ubiquitination may associate with changes in
catalytic activity, DNA binding activity, and/or stability of DNMT1 [15-18]. Recently,
acetylation of DNMT1 has been suggested in two global proteomics analysis [19, 20]. Silent
mating type information regulation 2 homolog 1 (SIRT1) is a type of Class III histone
deacetylases (HDACs), and has been reported to deacetylate the DNMT1 protein and alter its
activities [21]. Interestingly, SIRT1 deficiency resulted in the development of an autoimmune
syndrome in mice that manifests as a high titer of anti-nuclear antibody in serum,
immunoglobulin deposition in the kidney, and immune complex glomerulonephritis [22, 23].
6
the development of autoimmune diseases, such as SLE, although the underlying mechanistic
cues are poorly defined.
The purpose of this study was to investigate whether SIRT1 was involved in the
UVB-induced DNMT1 activity inhibition in SLE CD4+ T cells, and if so, what the underlying
mechanism is. We report here, for the first time, that UVB inhibits DNMT1 activity via
AhR-dependent downregulation of SIRT1 in CD4+ T cells from SLE patients.
2. Materials and methods 2.1. Subjects
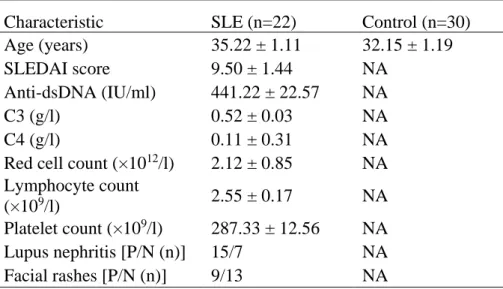
According to 1997 ACR revised criteria for classification of SLE [24] and SLE disease
activity index (SLEDAI, active disease was defined as an SLEDAI score ≥ 5), 22 newly
diagnosed active SLE patients (mean age: 35.22 ± 1.11 years) were recruited from the
outpatient department of Shanghai First People’s Hospital. Thirty age- and sex-matched
healthy controls (mean age: 32.15 ± 1.19 years) were recruited from the medical staff at
Shanghai First People’s Hospital. Relevant clinical information regarding the study subjects
is presented in Table 1. Because of the low number of T cells in most SLE patients and
limitations in the amount of blood collected per patient, not all experiments could be
conducted in each individual patient. This study was approved by the Human Ethics
Committee of Shanghai Jiaotong University, and written informed consent was obtained from
each subject.
2.2. Isolation, culture and treatment of CD4+ T cells
Blood samples (approximately 60 mL) were obtained from all participants. CD4+ T cells
were purified by negative selection using the RosetteSepTM Human CD4+ T Cell Enrichment
7
centrifugation (Histopaque; Sigma-Aldrich, St Louis, MO, USA). The purity of the CD4+ T
cells was evaluated by flow cytometry (purity ≥ 95%; BD Biosciences, New York, NY,
USA). The cells were then cultured in Xvivo 15 medium (Lonza, Walkersville, MD, USA)
supplemented with 10% human AB serum (Valley Biomedical, Winchester, VA, USA) at
37ºC with 5% CO2. Where indicated, the cells were treated with SRT1720
(Calbiochem-Novabiochem Corp., La Jolla, CA, USA) or (4-(3-Chloro-phenyl)-pyrimidin-2
-yl)-(4-trifluoromethyl-phenyl)-amine (VAF347, EMD Millipore, Billerica, MA, USA). UVB
treatment (50 mJ/cm2) of cells was performed as previously described by our laboratory [6].
2.3. Reverse-transcription PCR (RT-PCR) and quantitative real-time PCR (qRT-PCR)
analysis
Total RNA was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA). Reverse
transcription was performed using the PrimeScript RT-PCR kit (Takara Bio, Shiga, Japan).
qRT-PCR was performed on the Mx3000p real-time system (Stratagene, La Jolla, CA) using
SYBR Premix Ex Taq (Takara Bio). The amplification protocol included an initial
denaturation step at 95ºC for 30 seconds, followed by 40 successive cycles of 95ºC for 5
seconds and 60ºC for 20 seconds. The primers (Takara Bio) are presented in Table 2.
2.4. Western blotting analysis
CD4+ T cells were incubated with Complete Lysis-M (Roche Applied Science, Indianapolis,
IN, USA). Lysate protein concentration was measured using BCA Protein Assay kit (Pierce,
Rockford, IL, USA). Equal amounts of protein (20µg) were dissolved in NuPage LDS
Sample Buffer (Invitrogen, Carlsbad, CA, USA) and 10% NuPage Sample Reducing Agent
(Invitrogen). Lysates were boiled at 70ºC for 10 min and loaded and run on 4–12% NuPage
8
polyvinylidene fluoride membranes (Invitrogen) and blocked in 2% BSA in 0.1% Tween-20
(Sigma-Aldrich) and Tris-buffered saline. Membranes were probed with anti-SIRT1 rabbit
monoclonal IgG antibody (ab32441, Abcam, Cambridge, MA, USA), anti-AhR mouse
monoclonal IgG antibody (ab2769, Abcam) or anti-glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) rabbit IgG antibody (FL-335, Santa Cruz Biotechnology, CA, USA
USA) overnight at 4ºC. The secondary antibody used was anti-rabbit or anti-mouse
horseradish peroxidase-conjugated IgG antibody (Santa Cruz). Protein bands were detected
using the Western Breeze kit (Invitrogen).
2.5. Nuclear extraction and DNMT1 catalytic activity detection
Nuclear extracts were prepared by using EpiQuik™ Nuclear Extraction Kit I (Epigentek
Group Inc., Brooklyn, NY, USA). DNMT1 activity was calculated by using EpiQuik™ DNA
Methyltransferase Activity/Inhibition Assay Kit (Fluorometric) (Epigentek). DNMT1
catalytic activity was calculated by using the following formula:
DNMT activity (RFU/h/mg) = (No inhibitor RFU – blank RFU) × 1000/ Protein amount (μg)
× hour.
2.6. Transfection with SIRT1 or AhR -targeted specific small interference RNA
SiRNA targeted against AhR (si-AhR, s1200), SIRT1 (si-SIRT1, HSS177403) and siRNA
consisting of a scrambled sequence that would not lead to specific degradation of any cellular
message (si-control) were purchased from Ambion (Austin, TX, USA). CD4+ T cells
cultured in 24-well plates were incubated with mix from HiPerFect Transfection kit (Qiagen,
Courtaboeuf, France) containing 10 nM siRNA and 3.0 ml of HiPerFect reagent in 0.5 ml of
9
treated as indicated. siRNA transfection showed no effect on cell viability, as demonstrated
by microscopic examination (data not shown).
2.7. Chromatin Immunoprecipitation
ChIP assay of cultured CD4+T cells was performed using the EZ ChIP Chromatin
Immunoprecipitation Kit, Upstate Biotech (17-371, Merck Millipore). Briefly, 2×107 cells
were cross-linked with 1% formaldehyde, washed in PBS, and lysed in SDS lysis buffer.
Chromatin was fragmented by sonication and precleaned with protein A agarose. Samples
were incubated with anti-AhR (ab2769, Abcam) or unspecific mouse IgG antibodies (Santa
Cruz), and complexes were immunoprecipitated with protein A agarose. Protein-DNA
complexes were eluted from the antibodies with 1% SDS, 0.1 M NaHCO3, and DNA-protein
interactions were reversed by addition of 5 M NaCl and heating to 65 °C for 4 h. Proteins
were digested with proteinase K, and the remaining DNA was purified by phenol/chloroform
extraction. Site-specific PCR was carried out using 8 specific primer pairs (each covered
400bp) designed using Primer premier5.0 software, amplifying the SIRT1 promoter region
between 2kb upstream of start site TSS and 1kb downstream. Each ChIP experiment was
carried out at least three times with similar results. The primers (Takara Bio) are presented in
Table 3.
2.8. Statistical analysis
All data are presented as the means ± SEM and were analyzed by the independent-samples t
-test or one-way ANOVA followed by Dunnett’s multiple comparisons post-test. All analyses
10 3. Results
3.1 DNMT1 activity was modulated by SIRT1 expression in CD4+ T cells
SIRT1 appeared to alter DNMT1 activity in several different cell lines, such as HeLa and
293T cells [21], however, whether SIRT1 was essentially involved in the modulation of
DNMT1 activity in CD4+ T cells was unknown. Fig. 1A showed that the SIRT1 expression
was knocked down by using SIRT1 siRNA in CD4+ T cells. Forty-eight hours later, DNMT1
activity was detected and markedly inhibited compared to si-control transfected CD4+ T cells
(Fig. 1B). Next, we examined whether SIRT1 expression upregulation could induce the
DNMT1 activity in CD4+ T cells. CD4+ T cells were stimulated with SRT1720, the
established SIRT1 activator, at the concentration of 10 and 20 μM. The mRNA and protein
expression of SIRT1 were measured 6 or 24 hours after treatment respectively. Data showed
that the mRNA and protein expression level of SIRT1 was increased by SRT1720 in a
concentration-dependent manner (Fig. 1C and D). Then as expected, the DNMT1 activity
was induced by SRT1720 in CD4+ T cells in a concentration-dependent manner (Fig. 1E). To
rule out the possibility that increased DNMT1 activity in CD4+ T cells was driven by
SRT1720 by mechanisms other than activating SIRT1, we examined the inductive effect on
DNMT1 activity by SRT1720 from si-control- and si-SIRT1-transfected CD4+ T cells. As
depicted in Fig. 1F, the inductive effect on DNMT1 activity by SRT1720 was cancelled in
the si-SIRT1 condition. Using these in vitro approaches, we supposed that SIRT1 expression
could modulate the DNMT1 activity in CD4+ T cells.
3.2 UVB inhibited DNMT1 activity via decreasing SIRT1 expression in CD4+ T cells
11
Our previous work demonstrated that UVB downregulated DNMT1 activity in a
dosage-dependent manner in lupus CD4+ T cells, but not in CD4+ T cells from healthy controls[6].
Given the critical role of SIRT1 expression for the DNMT1 activity modulation, we first
tested whether UVB could affect SIRT1 expression in CD4+ T cells. CD4+ T cells from
healthy controls and SLE patients were exposed to 50 mJ/cm2 UVB irritation as previous
published [6]. We showed that both mRNA and protein expression level of SIRT1 were
inhibited by UVB treatment in lupus CD4+ T cells. By contrast, the SIRT1expression in
normal CD4+ T cells was comparable prior to and after UVB stimulation (Fig. 2A, B and C).
We next assessed whether addition of SRT1720 could restore UVB-mediated DNMT1
activity inhibition in lupus CD4+ T cells. CD4+ T cells form SLE patients were pretreated
with 20 μM SRT1720 for 60 min, and then exposed to 50 mJ/cm2 UVB irradiation. As
depicted in Fig. 2D, UVB-inhibited DNMT1 activity was reversed by SRT1720 in lupus
CD4+ T cells. However, the reversal effect of SRT1720 was not detected in
si-SIRT1-transfected lupus CD4+ T cells with UVB stimulation. Together, these findings demonstrated
UVB downregulated SIRT1 expression and subsequently inhibited DNMT1 activity in lupus
CD4+ T cells.
3.3 SIRT1 expression was directly and negatively regulated by AhR
AhR has been reported to link UVB with SLE [25] and therefore, we sought to determine
whether AhR activation was involved in the regulation of SIRT1 expression in CD4+ T cells.
Indeed, we observed that normal CD4+ T cells stimulated with 10 and 50 nM VAF347, the
specific AhR agonist, strongly inhibited SIRT1 mRNA and protein in a
concentration-dependent manner (Fig. 3A and B). We suspected that the SIRT1 gene promoter might
contain a functional AhR binding site, and the downregulation of SIRT1 resulted from
12
antibody against AhR in normal CD4+ T cells. A 16-fold stronger ChIP signal was observed
using primers that amplify a product at the site about 1.6 kb upstream from the translation
start site of the SIRT1 promoter while Control Ab ChIP assays did not result in significant
enrichment (Fig. 3C). These data indicated that there might be a promoter repressor element
located at the distal sequence upstream of the SIRT1 gene.
3.4 AhR activation was involved in the UVB-induced SIRT1/DNMT1 axis inhibition in
CD4+ T cells from SLE patients
We first investigated the potential of UVB to activate AhR using CD4+ T cells from healthy
controls and SLE patients. Six hours after 50 mJ/cm2 UVB irritation, an increase in
expression of typical AhR-responsive genes CYP1A1 was observed in both UVB–treated
normal and lupus CD4+ T cells (Fig. 4A), while expression of CYP2E1 (not an AhR target)
was unaffected (data not shown). Results also showed that SLE patients expressed higher
levels of CYP1A1 transcripts following UVB-induced AhR activation, as compared with
healthy controls (Fig. 4A). To confirm the regulatory role of the AhR signaling pathway in
the UVB–mediated inhibition of SIRT1/DNMT1 axis, we studied the UVB response after
siRNA-mediated knockdown of AhR in lupus CD4+ T cells. As shown in Fig. 4B, the protein
expression of AhR was almost knocked down by using AhR-targeted siRNA. Concomitantly,
AhR deficiency abrogated the inhibition of UVB-mediated SIRT1 mRNA and protein
expression (Fig. 4C and D) and DNMT1 activity in lupus CD4+ T cells (Fig. 4E).
Collectively, these data suggested that AhR activation might play a crucial and nonredundant
role in UVB-induced SIRT1/DNMT1 axis inhibition in lupus CD4+ T cells.
13
Of best-known inducers that trigger SLE flares, UV light is the strongest. We supposed, as
suggested by others [26], that UV exposure aggravated lupus via enhancing DNA
hypomethylation. Our previous study also reported that UVB inhibited DNMT1 activity in
lupus CD4+ T cells [6]. In the present study, we demonstrated that UVB suppressed SIRT1
expression via activating AhR, and subsequently inhibited DNMT1 activity in CD4+ T cells
from SLE patients.
Given its fundamental importance, the key DNA methyltransferase enzyme, DNMT1, is
tightly regulated in mammalian cells. Peng et al.[21] previously showed that DNMT1 was an
acetylated protein, and its DNA methyltransferase activity was increased by SIRT1 through
deacetylation. Our results fit well with their observation. In the current study, deletion of
SIRT1 gene expression in vitro by SIRT1-targeted siRNA treatment of normal CD4+ T cells
resulted in a dramatically decreased DNMT1 activity. Because of limitation in the amount of
human blood sample and massive cell death after SIRT1-expressing retrovirus transduction
(data not shown) we treated CD4+ T cells by SRT1720, a well-accepted SIRT1 activator, as
an alternative method to upregulate SIRT1 expression in normal CD4+ T cells. In line with
Jia et al. [27], our data showed that SIRT1 mRNA and protein expression were induced by
SRT1720. We next demonstrated that DNMT1 activity was also increased by SRT1720 in a
concentration-dependent manner. The inductive effect of SRT1720 on DNMT1 activity was
abolished in si-SIRT1-transfected CD4+ T cells which excluded the possibility that SRT1720
increased DNMT1 activity through other signal pathways other than SIRT1 upregulation.
However, the possible mechanisms of SIRT1-stimulation on DNMT1 activity upregulation
other than deacetylation are not ruled out by our work.
Several lines of evidence indicated that SIRT1 negatively regulated T cells activation,
and loss of SIRT1 resulted in a breakdown of CD4+ T cells tolerance [23, 28, 29]. By
14
active lupus CD4+ T cells compared with controls. In this study, we observed that the
expression of SIRT1 was comparable between normal and lupus CD4+ T cells. These
controversial data indicated that further investigations were needed to elucidate the exact role
of SIRT1 in the pathogenesis of SLE. Consistent with previous studies [31, 32], we showed
that the expression of SIRT1 was reduced with UVB irritation in CD4+ T cells. Interestingly,
the SIRT1 expression was remarkably inhibited in lupus CD4+ T cells whereas only slightly
decreased in normal CD4+ T cells after UVB exposure. These data likely reflected intrinsic
differences of SIRT1 response between normal and lupus CD4+ T cells.
We speculated that AhR might paly roles in the modulation of SIRT1 expression and
the different SIRT1 response to UVB irritation between normal and lupus CD4+ T cells,
because AhR could be activated by UVB [33], and AhR has been reported to link UVB with
SLE [25]. As expected, SIRT1 expression was downregulated via AhR avctivation by
VAF347. By using ChIP assay we demonstrated a putative functional AhR binding site might
exist at the distal sequence upstream of the SIRT1 gene promoter. Collectively, results from
these in vivo studies strongly suggested that AhR could regulate the SIRT1 expression in
CD4+ T cells. Moreover, we showed that UVB-induced AhR response in lupus CD4+ T cells
was much stronger than that in normal CD4+ T cells. These results were also supported by
Gorochov et al. [12] that a more significant AhR activation was observed in the peripheral
blood of active SLE patients. Gorochov et al. [12] suggested that such "AhR hyper
responsive state" was likely to be associated with increased numbers of circulating IL-17
and/or IL-22-secreting cells in SLE patients. These observations need further investigation
and will be addressed in our future studies. Finally, our data showed that UVB-inhibited
SIRT1 expression was markedly blocked in si-AhR-transfected lupus CD4+ T cells. Taken
together, our findings indicated that AhR mediated UVB-inhibited SIRT1 in CD4+ T cells
15
In conclusion, our study demonstrated that UVB strongly activated AhR, and
UVB-activated AhR downregulated the expression of SIRT1 via binding to the SIRT1 promoter.
Subsequently, decreased SIRT1 resulted in the DNMT1 activity inhibition in CD4+ T cells
from SLE patients (Fig. 5). Our data also indicated that AhR and/or SIRT1 might be potential
therapeutic targets of SLE.
Funding source: National Natural Science Foundation of China (grant nos. 81402587 and
81573031).
The authors have no conflict of interest to declare.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (grant nos.
81402587 and 81573031).
References
[1] Hewagama A, Richardson B. The genetics and epigenetics of autoimmune diseases.
Journal of autoimmunity 2009; 33: 3-11.
[2] Richardson BC, Liebling MR, Hudson JL. CD4+ cells treated with DNA methylation
inhibitors induce autologous B cell differentiation. Clinical immunology and
immunopathology 1990; 55: 368-81.
[3] Zopf S, Ocker M, Neureiter D, et al. Inhibition of DNA methyltransferase activity and
expression by treatment with the pan-deacetylase inhibitor panobinostat in hepatocellular
carcinoma cell lines. BMC cancer 2012; 12: 386.
[4] Anderson RM, Bosch JA, Goll MG, et al. Loss of Dnmt1 catalytic activity reveals
multiple roles for DNA methylation during pancreas development and regeneration.
16
[5] Romanek-Piva K, Galczynski K, Adamiak-Godlewska A, et al. DNA methylation and
DNA methyltransferase (DNMT1) activity pattern in endometrial carcinoma. Ginekologia
polska 2016; 87: 6-10.
[6] Wu Z, Li X, Qin H, et al. Ultraviolet B enhances DNA hypomethylation of CD4+ T cells
in systemic lupus erythematosus via inhibiting DNMT1 catalytic activity. Journal of
dermatological science 2013; 71: 167-73.
[7] Hahn ME. Aryl hydrocarbon receptors: diversity and evolution. Chemico-biological
interactions 2002; 141: 131-60.
[8] Stockinger B, Di Meglio P, Gialitakis M, et al. The aryl hydrocarbon receptor:
multitasking in the immune system. Annu Rev Immunol 2014; 32: 403-32.
[9] Nguyen NT, Hanieh H, Nakahama T, et al. The roles of aryl hydrocarbon receptor in
immune responses. International immunology 2013; 25: 335-43.
[10] Singh NP, Singh UP, Singh B, et al. Activation of aryl hydrocarbon receptor (AhR) leads
to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of
experimental colitis. PloS one 2011; 6: e23522.
[11] Esser C, Bargen I, Weighardt H, et al. Functions of the aryl hydrocarbon receptor in the
skin. Seminars in immunopathology 2013; 35: 677-91.
[12] Dorgham K, Amoura Z, Parizot C, et al. Ultraviolet light converts propranolol, a
nonselective beta-blocker and potential lupus-inducing drug, into a proinflammatory AhR
ligand. European journal of immunology 2015; 45: 3174-87.
[13] Dhe-Paganon S, Syeda F, Park L. DNA methyl transferase 1: regulatory mechanisms and
implications in health and disease. International journal of biochemistry and molecular
17
[14] Kar S, Deb M, Sengupta D, et al. An insight into the various regulatory mechanisms
modulating human DNA methyltransferase 1 stability and function. Epigenetics 2012; 7:
994-1007.
[15] Esteve PO, Chin HG, Benner J, et al. Regulation of DNMT1 stability through
SET7-mediated lysine methylation in mammalian cells. Proceedings of the National Academy of
Sciences of the United States of America 2009; 106: 5076-81.
[16] Lee B, Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. The
Biochemical journal 2009; 421: 449-61.
[17] Sugiyama Y, Hatano N, Sueyoshi N, et al. The DNA-binding activity of mouse DNA
methyltransferase 1 is regulated by phosphorylation with casein kinase 1delta/epsilon. The
Biochemical journal 2010; 427: 489-97.
[18] Zhou Q, Agoston AT, Atadja P, et al. Inhibition of histone deacetylases promotes
ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast
cancer cells. Molecular cancer research : MCR 2008; 6: 873-83.
[19] Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and
co-regulates major cellular functions. Science (New York, N.Y.) 2009; 325: 834-40.
[20] Kim SC, Sprung R, Chen Y, et al. Substrate and functional diversity of lysine acetylation
revealed by a proteomics survey. Molecular cell 2006; 23: 607-18.
[21] Peng L, Yuan Z, Ling H, et al. SIRT1 deacetylates the DNA methyltransferase 1
(DNMT1) protein and alters its activities. Molecular and cellular biology 2011; 31: 4720-34.
[22] Sequeira J, Boily G, Bazinet S, et al. sirt1-null mice develop an autoimmune-like
condition. Experimental cell research 2008; 314: 3069-74.
[23] Zhang J, Lee SM, Shannon S, et al. The type III histone deacetylase Sirt1 is essential for
maintenance of T cell tolerance in mice. The Journal of clinical investigation 2009; 119:
18
[24] Passas CM, Wong RL, Peterson M, et al. A comparison of the specificity of the 1971
and 1982 American Rheumatism Association criteria for the classification of systemic lupus
erythematosus. Arthritis and rheumatism 1985; 28: 620-3.
[25] Esser C, Rannug A, Stockinger B. The aryl hydrocarbon receptor in immunity. Trends in
immunology 2009; 30: 447-54.
[26] Zhu X, Li F, Yang B, et al. Effects of ultraviolet B exposure on DNA methylation in
patients with systemic lupus erythematosus. Experimental and therapeutic medicine 2013; 5:
1219-25.
[27] Jia Y, Zheng Z, Wang Y, et al. SIRT1 is a regulator in high glucose-induced
inflammatory response in RAW264.7 cells. PloS one 2015; 10: e0120849.
[28] Zou T, Yang Y, Xia F, et al. Resveratrol Inhibits CD4+ T cell activation by enhancing
the expression and activity of Sirt1. PloS one 2013; 8: e75139.
[29] Kong S, Yeung P, Fang D. The class III histone deacetylase sirtuin 1 in immune
suppression and its therapeutic potential in rheumatoid arthritis. Journal of genetics and
genomics = Yi chuan xue bao 2013; 40: 347-54.
[30] Hu N, Qiu X, Luo Y, et al. Abnormal histone modification patterns in lupus CD4+ T
cells. The Journal of rheumatology 2008; 35: 804-10.
[31] Benavente CA, Schnell SA, Jacobson EL. Effects of niacin restriction on sirtuin and
PARP responses to photodamage in human skin. PloS one 2012; 7: e42276.
[32] Chou WW, Chen KC, Wang YS, et al. The role of SIRT1/AKT/ERK pathway in
ultraviolet B induced damage on human retinal pigment epithelial cells. Toxicology in vitro :
an international journal published in association with BIBRA 2013; 27: 1728-36.
[33] Jux B, Kadow S, Luecke S, et al. The aryl hydrocarbon receptor mediates UVB
19 Figure Legends
Figure 1. DNMT1 activity was modulated by SIRT1 expression in CD4+ T cells.
(A) Western blotting analysis for the effect of si-SIRT1 transfection. (B) Normal CD4+ T
cells or si-SIRT1-transfected normal CD4+ T cells were exposed to UVB (50 mJ/cm2).
Twenty-four hours following UVB irradiation, the DNMT1 activity was detected. (C)
Normal CD4+ T cells were treated with SRT1720 (10 and 20 μM) for 6 h for qRT-PCR
analysis. SIRT1 mRNA levels were normalized to GAPDH mRNA levels. (D) Normal CD4+
T cells were treated with SRT1720 (10 and 20 μM) for 24 h for Western blotting analysis.
SIRT1 protein levels were normalized for GAPDH protein levels using ImageJ software. (E)
Normal CD4+ T cells were treated with SRT1720 (10 and 20 μM) for 24 h for DNMT1
activity detection. (F) si-control- or si-SIRT1-transfected normal CD4+ T cells were
incubated with SRT1720 (20 μM) for 24 h for DNMT1 activity detection. Results are
expressed as mean ± SEM (n = 3) of one representative experiment of at least three. **P <
0.01, ***P < 0.001. ns, not significant; unpaired Student's t test (B) or one-way ANOVA
followed by Dunnett multiple comparisons posttest (C, E and F).
Figure 2. UVB inhibited DNMT1 activity via decreasing SIRT1 expression in CD4+ T
cells from SLE patients.
(A) CD4+ T cells were isolated from SLE patients (n=20) and healthy controls (n=20). CD4+
T cells form SLE patients (n=10) and healthy controls (n=10) were exposed to UVB (50
mJ/cm2). Six hours following UVB irradiation, the mRNA expression level of SIRT1 was
measured by qRT-PCR. SIRT1 mRNA levels were normalized to GAPDH mRNA levels.
Each dot represents an untreated individual healthy control. Each square represents a treated
20
inverted triangle represents a treated individual SLE patient. (B) Western blotting and (C)
quantitative analysis. CD4+ T cells form SLE patients and healthy controls were exposed to
UVB (50 mJ/cm2), and 24 hours following UVB irradiation, the protein expression level of
SIRT1 was measured. SIRT1 protein levels were normalized to GAPDH protein levels using
ImageJ software. (D) Si-control- or si-SIRT1-transfected lupus CD4+ T cells were exposed to
UVB (50 mJ/cm2) with or without 60 min pretreatment with SRT1720 (20 μM). Twenty-four
hours following UVB irradiation, the DNMT1 activity was detected. Results are expressed as
mean ± SEM (n = 3) of one representative experiment of at least three. (B-D) **P < 0.01,
***P < 0.001; ns, not significant, one-way ANOVA followed by Dunnett multiple
comparisons posttest. (A-D)
Figure 3. SIRT1 expression was directly and negatively regulated by AhR .
(A) Normal CD4+ T cells were treated with VAF347 (10 and 50 nM) for 6 h for qRT-PCR
analysis. SIRT1 mRNA levels were normalized to GAPDH mRNA levels. (B) Normal CD4+
T cells were treated with VAF347 (10 and 50 nM) for 24 h for Western blotting analysis.
SIRT1 protein levels were normalized for GAPDH protein levels using ImageJ software. (C)
ChIP analysis. ChIP using an AhR Ab or IgG control Ab revealed binding of AhR to the
SIRT1 promoter in a distal upstream region in normal CD4+ T cells. Results are expressed as
mean ± SEM (n = 3) of one representative experiment of at least three. (A-C) *P < 0.01,
***P < 0.001; one-way ANOVA followed by Dunnett multiple comparisons posttest. (A and
C)
Figure 4. AhR activation was involved in the UVB-induced SIRT1/DNMT1 axis
21
(A) CD4+ T cells form SLE patients (n=10) and healthy controls (n=10) were exposed to
UVB (50 mJ/cm2). Six hours following UVB irradiation, the mRNA expression level of
CYP1A1 was measured by qRT-PCR. CYP1A1 mRNA levels were normalized to GAPDH
mRNA levels. Each dot represents an individual healthy control. Each square represents an
individual healthy control. (B) Western blotting analysis for the effect of si-AhR transfection.
(C) Si-control- or si-AhR-transfected lupus CD4+ T cells were exposed to UVB (50 mJ/cm2).
Six hours following UVB irradiation, the mRNA expression level of SIRT1 was measured by
qRT-PCR. SIRT1 mRNA levels were normalized to GAPDH mRNA levels. (D) Western
blotting and (E) quantitative analysis. Si-control- or si-AhR-transfected lupus CD4+ T cells
were exposed to UVB (50 mJ/cm2), and 24 hours following UVB irradiation, the protein
expression level of SIRT1 was measured. SIRT1 protein levels were normalized to GAPDH
protein levels using ImageJ software. (F) Si-control- or si-AhR-transfected lupus CD4+ T
cells were exposed to UVB (50 mJ/cm2), and 24 hours following UVB irradiation, the
DNMT1 activity was detected. Results are expressed as mean ± SEM (n = 3) of one
representative experiment of at least three. (B-F) *P < 0.01, ***P < 0.001, ****P < 0.0001;
one-way ANOVA followed by Dunnett multiple comparisons posttest. (A, C, E and F)
Figure 5. A working model of the molecular mechanism of UVB inhibition of DNMT1
activity in CD4+ T cells from SLE patients.
First, UVB strongly activates AhR in lupus CD4+ T cells. Then, the activated AhR binds to
the SIRT1 promoter in a distal upstream region, leading to the downregulation of SIRT1
expression. SIRT1 suppression then has a negative effect on the DNMT1 activity in lupus
CD4+ T cells. Red arrows show effects of experimental manipulation; black T-bar,
suppression; black arrow, stimulatory activity; filled circle, putative AhR binding site site in
Figure 1
B
E
Figure 2
D
C
Figure 3
Figure 4
A
F
E
22
Table 1. Clinical and laboratory characteristics of the subjects
Characteristic SLE (n=22) Control (n=30)
SLEDAI = Systemic Lupus Erythematosus Disease Activity Index; Values are presented as mean ± SEM, except where indicated otherwise. P/N: positive/negative.
Table 2. Sequences of primers used for qRT-PCR
23 Table 3. Sequences of primers used for ChIP
Primer name Sequence (5'-3')
Primer1 FW TTTTGAGACGGAGTTTCGC
Primer1 RV CAGAAGGCTGAGGCAGGA
Primer2 FW ACTTCCGACTTCAGGTGATC
Primer2 RV GCTAAGGTCCTATCTACATCCA
Primer3 FW GCCTAAAGTCACGCAGGTA
Primer3 RV CCAGTGTTTGTTATGGCATCT
Primer4 FW AAACGGCTAGATAGCTCACG
Primer4 RV GCAGAATGGGTTTGTTGG
Primer5 FW CAGAACGACTATCCAACGTA
Primer5 RV TGACCTCAAATCACTACCG
Primer6 FW TACACGCTCGCCACAAAG
Primer6 RV CCAGACCACAACACTACGG
Primer7 FW CCCCAGAGCGTGAGGTGC
Primer7 RV AGTTGTCGGCCAGCGGTG
Primer8 FW CCTACTGGCCTGAGGTTG