Differential diurnal expression of rice catalase genes: the
5
%
-flanking region of
CatA
is not sufficient for circadian control
Masao Iwamoto, Hiromi Higo
1, Kenichi Higo *
Department of Genetic Resources II,National Institute of Agrobiological Resources,Tsukuba,Ibaraki305-8602,Japan
Received 15 June 1999; received in revised form 27 August 1999; accepted 23 September 1999
Abstract
Three rice catalase genes (CatA,CatB,CatC) are expressed in a growth- and tissue-specific manner. In seedlings, theCatA,
CatBandCatCgenes were highly expressed in the leaf sheath, root and leaf blade, respectively. The expression of theCatAgene in the leaf sheath was modulated by a circadian rhythm with the maximum late in the light period. On the other hand, diurnal oscillations ofCatCexpression were detected in the leaf blade when plants were grown in the dark. Theb-glucuronidase (GUS) gene driven by the 5%-flanking region ofCatAwas expressed with diurnal fluctuations, the pattern of which is different from that of the endogenousCatAmRNA, both at the pre-mRNA and at the mRNA level in transgenic plants. These results suggest that the circadian rhythmic expression of CatA is due to a transcriptional or post-transcriptional event such as modulation of pre-mRNA stability and requires some other region(s), in addition to the promoter region, exon-1 and intron-1. © 2000 Elsevier Science Ireland Ltd. All rights reserved.
Keywords:Catalase; Circadian rhythm; GUS;Oryza sati6aL.; Rice; Tissue-specific expression
www.elsevier.com/locate/plantsci
1. Introduction
Catalase (H2O2:H2O2 oxidoreductase, EC
1.11.1.6) is one of the enzymes necessary for de-composing the toxic intermediate of oxygen metabolism, H2O2, into oxygen and water. In
higher plants, catalases scavenge the H2O2
pro-duced, for example, during b-oxidation of fatty acids in seeds or during photorespiration in leaves. Plant catalase genes comprise a small gene fam-ily. Three catalase genes have been identified in maize (Zea mays) [1 – 3], Arabidopsis thaliana [4] and tobacco (Nicotiana plumbaginifolia) [5] and pumpkin (Cucurbitasp.) [6], and were shown to be
differentially expressed [4,6 – 9]. In maize, for ex-ample, one of the three maize genes, Cat1, is expressed in the root, epicotyl and scutellum, whereasCat2 transcript accumulates to high levels in the scutellum and leaf, and strong expression of
Cat3 is detected in the epicotyl and leaf [8]. It has been reported that the level of Cat3 mRNA is under the control of a circadian rhythm [10].
Rice (Oryza sati6a) also contains three catalase genes (CatA, CatB, CatC) ([11,12], Higo et al. in preparation). To understand the mode of expres-sion of the three catalase genes in rice, we per-formed Northern analysis, using gene specific probes, of total RNAs from various tissues and from plants harvested at different times under light/dark (LD), continuous light (LL) or continu-ous dark (DD) conditions. Furthermore, to eluci-date whether the 5%-flanking region ofCatA, a rice ortholog ofCat3, is sufficient to confer a circadian expression, we examined transgenic rice plants, which contain the b-glucuronidase (GUS) gene driven by the 5%-flanking region of CatA, by * Corresponding author. Present address: Department of
Molecu-lar Genetics, National Institute of Agrobiological Resources, Tsukuba, Ibaraki 305-8602, Japan. Tel.:+81-298-387011; fax:+ 81-298-387492.
E-mail address:[email protected] (K. Higo)
1Present address: Department of Molecular Genetics, National
Institute of Agrobiological Resources, Tsukuba, Ibaraki 305-8602, Japan
Northern blot and reverse transcription-poly-merase chain reaction (RT-PCR) analyses.
2. Materials and methods
2.1. Plant materials and growth conditions
Rice plants (O. sati6a cv. Nipponbare) were grown for 5 weeks (seedlings at 4.5 leaf-age) or for 3 months (mature plants at flowering stage) in a green house. For detecting circadian rhythmic ex-pression, rice seedlings were grown in a growth chamber under a light-dark (LD) cycle regimen (16 h of light and 8 h of dark) at 28°C for 3 weeks (six leaf-age), and then some of them were trans-ferred to LL or DD conditions at the end of the dark period. White light (160mmol m−2s−1) was
provided by white fluorescent lights. Transgenic rice plants, which were derived from O. sati6a cv.
Kinuhikari, were produced by Drs H. Akagi and T. Fujimura. Protoplasts were prepared and elec-troporated in the presence of pBI221 (Clontech) containing the GUS gene fused to the 5%-flanking
region (−1564 to +342, the cDNA start site of
CatA being designated as +1) of CatAinstead of the CaMV35S promoter, and pUC19-HPT con-taining the hygromycin phosphotransferase (HPT) gene linked to the CaMV35S promoter, and regen-erated according to methods previously described [13].
2.2. RNA extraction and Northern blot analysis
Total RNA was extracted from the leaf blades, leaf sheaths, roots of both seedlings and mature plants, and flower sampled at the stages from −6 to +1 days after pollination (DAP) using RNeasy™(QIAGEN) according to the instruc-tions provided by the manufacturer. For detecting circadian rhythmic expression, leaf blades and leaf sheaths were harvested every 4 h for 52 h, 2 h after their transferal to LL or DD conditions, and stored at −80°C until RNA extraction. Total RNAs (20 mg) were electrophoresed on a 1% agarose gel containing formaldehyde and then transferred to a Hybond™-N+ membrane
(Amer-sham). Catalase gene-specific probes, which cover the 3%untranslated region of each gene, were
pro-duced by polymerase chain reaction (PCR). The probes for CatA (305 bp), CatB (207 bp) and
CatC (186 bp) were amplified using A-F (5%-TCA
TCT CTT GTT AAT TAA TTG GAG TAC TAC-3%) and A-R (5%-GAA GTG ATA ATT TAA
ATA CTT AAT AGT AAT-3%), B-F (5%-CCA GTG GTG GTG CTA TGT TGG ACA GTC AAA-3%) and B-R (5%-AAA CAA GTT CAA ACA
TAT AGC ACC CAC TGT-3%), and C-F (5%-GAG ATC GAT CAG CGT TGC AAT CTG CTT CAG-3%) and C-R (5%-CTT CAG GTT ACA GAT TAT TAC ATG ATG GTG-3%) primers,
respec-tively. The nucleotide sequences of the three cata-lase genes have been deposited in the DDBJ/EMBL/GenBank databases under the ac-cession numbers of D29966 (CatA), D64013 (CatB) and D86611 (CatC) (see [11,12]). GUS gene-specific probe (952 bp), which corresponds to a part of the coding region, was produced by PCR using G-F (5%-AAT TGA TCA GCG TTG GTG
G-3%) and G-R (5%-GAT GCC ATG TTC ATC TGC C-3%) primers. Radiolabeled DNA probes
were obtained by the random-priming method [14], except for the CatC-specific probe which was radiolabeled by PCR using the C-F and C-R primers. The membrane was hybridized with one of the probes at 42°C in a solution containing 50% formamide, 6×SSC, 5×Denhardt’s solution, 10% dextran sulfate, 0.1% SDS and 0.1 mg/ml single-stranded salmon sperm DNA for 1 day. The membrane was washed in 6×SSC for 60 min at 65°C, then visualized by autoradiography at −
80°C.
Relative levels ofCatA and CatC mRNAs were calculated from the intensity of radioisotopic sig-nals on X-ray films using the NIH image 1.61 program. The most abundant signal was defined as 1.00 and other abundances were shown relative to the maximum.
2.3. RT-PCR
Total RNA extracted from the leaf sheath of transgenic rice plants was treated with DNase I (amplification grade, Gibco-BRL) according to the instructions provided by the manufacturer. RT-PCR was performed using the Superscript™One-Step™RT-PCR system (Gibco-BRL). The DNase I-treated RNA (250 ng) was used as a template, and RT-PCR amplification reaction mixtures con-tained 0.2 mM of each dNTP, 1.2 mM MgSO4, 0.4
vol-ume of 25 ml. For amplification of the fragment derived from theCatA pre-mRNA, A-F2 (5%-GGC AGA AGG CGA CGA TAC-3%) and A-R2 (5%
-GTT GTG CTA AAA TGG TGT TAA TCA TGT G-3%) primers were prepared. A-F3 (5%-CAT
GGC TGG TTG ATT CAG C-3%) and G-R2
(5%-CGT CGG TAA TCA CCA TTC C-3%) primers were used to detect the presence of the chimeric GUS pre-mRNA. cDNA synthesis was achieved at 47°C for 40 min, then 94°C for 2 min, and PCR amplification was performed for 27 cy-cles of 1 min at 94°C, 2 min at 50°C and 2 min at 72°C using a Perkin-Elmer/Cetus DNA thermal cycler model PJ1000NPT. The RT-PCR products were fractionated on a 1.5% agarose gel and visu-alized by ethidium bromide staining.
3. Results
3.1. Differential expression of rice catalase genes during plant growth
Total RNAs of seedlings (4.5 leaf-age) and ma-ture plants of rice (O. sati6a cv. Nipponbare) were
extracted and used for Northern analysis. In seedlings, strong expression of CatA, CatB and
CatCwas detected in the leaf sheath, root and leaf blade, respectively (Fig. 1). Only weak expression of CatA was detected in the leaf blade, and of
CatB and CatC in the leaf sheath. The expression patterns of catalase genes in mature plants were, however, different from those in the seedlings. The
CatA gene was expressed in the leaf blade while suppressed in the leaf sheath, a strong expression
being observed in the flowers sampled at the stage from −6 to +1 DAP. The CatB showed weak expression in the leaf blade, leaf sheath, root and flower. The CatC mRNA was very abundant in the leaf blade, indicating that the tissue-specificity of CatC expression is maintained during the growth from seedling to mature plant.
3.2. Diurnal oscillations of the catalase gene transcripts
To examine whether the expression of the rice catalase genes is stable during the day and/or night, total RNAs were extracted from the shoot (a mixture of the leaf blade and leaf sheath) and root of rice seedlings, which were grown under an LD photoperiod and sampled at different time points. Northern analysis was performed using the shoot RNA for CatA and CatC, and the root RNA for CatB expression. These preliminary re-sults revealed that the accumulation of both CatA
and CatC mRNAs was variable among sampling points and that there was no obvious change in the CatB mRNA abundance.
It is known that the expression of some genes in plants shows diurnal fluctuations depending on the periodicity of light and dark in a day [10,15,16]. To examine whether the expression of CatA and
CatC is regulated by a circadian rhythm, seedlings grown under LD conditions and those transferred from LD to LL or DD were sampled every 4 h for 52 h. Total RNAs extracted from the leaf blade and leaf sheath were used for Northern analysis to examine patterns of the mRNA abundance. The
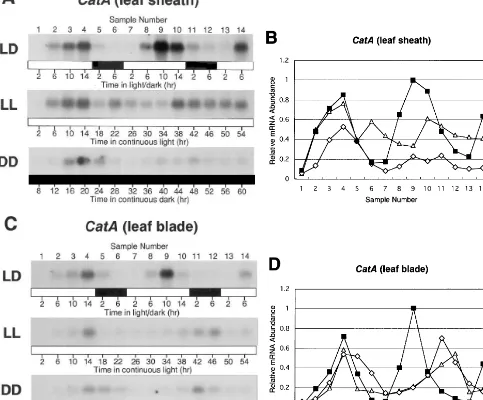
CatA transcript in the leaf sheath showed diurnal oscillations with the peak late in the light period (10 – 14 h after the dark/light transition) and with the trough late in the dark period and early in the light period (6 – 10 h after the light/dark transition) under LD conditions (Fig. 2A and B). A weak but similar pattern was observed with the leaf blade (Fig. 2C and D). Although diurnal oscillations of the CatA mRNA abundance were also seen under LL and DD conditions, a damping of fluctuations and a delayed second peak were observed (Fig. 2B and D). Damping of the CatAmRNA oscillations was more evident in the leaf sheath than leaf blade. Changes in diurnal oscillations under LL or DD conditions have been reported for the circa-dian-regulated catalase genes in tobacco, maize and Arabidopsis [9,10,17]. The abundance of 17S Fig. 1. Tissue-specific expression of three catalase genes in
Fig. 2. Circadian oscillations in CatAmRNA abundance. Rice seedlings were grown under LD, LL or DD conditions, and Northern blot analyses ofCatAmRNA in the leaf sheath (A) and leaf blade (C) were performed usingCatA-specific probe. Dark and light periods are indicated by black and white bars, respectively, below the figure. The sample number and the time of sample harvest in hours (hr) are indicated above and below the figure, respectively. Relative levels ofCatAmRNA in the leaf sheath (B) and leaf blade (D) were calculated from the intensity of radioisotopic signals in A and C, respectively.
rRNA, examined as a control in the leaf blade and leaf sheath, indicated a normal high expression level of this gene and no diurnal oscillations dur-ing the experimental period (data not shown).
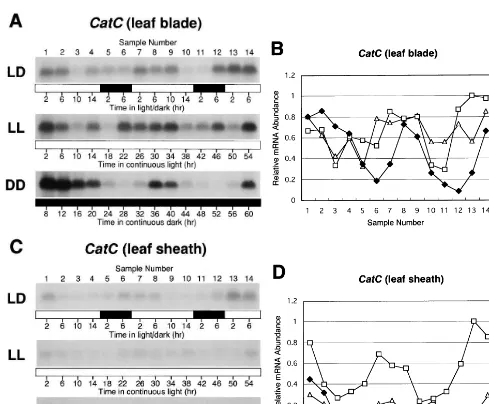
The fluctuations of the CatC transcript level in the leaf blade showed a pattern with a peak early in the light period and with a trough late in the light period and early in the dark period under LD conditions, but there were no obvious diurnal oscillations under LD and LL conditions (Fig. 3A and B). In contrast, daily fluctuations with a pat-tern of 24 h over a subjective 2 day period were observed under DD conditions. In the leaf sheath,
littleCatC mRNA accumulated, but daily fluctua-tions of theCatC mRNA were not detected under DD conditions (Fig. 3C and D).
3.3. Expression of the GUS gene dri6en by the
CatA promoter
The accumulation of CatA mRNA showed ro-bust daily oscillations under LD conditions (Fig. 2). The RNA extracted from the transgenic rice plants carrying a 1.9-kb DNA fragment contain-ing the 5%-flanking region, exon-1, intron-1 and a
reporter gene was used for Northern analysis to examine whether this region confers diurnal oscil-lations of the CatA mRNA. The chimeric GUS mRNA was abundant late in the light period as was the CatA mRNA, but showed daily fluctua-tions with an additional weak peak late in the dark period (Fig. 4). In contrast to the GUS mRNA, the diurnal oscillations in the endogenous level ofCatA mRNA seen in the transgenic plants are identical to those of CatA mRNA in non-transgenic rice in Fig. 2 (Fig. 4). These results indicate that the 1.9-kb fragment is not sufficient for the circadian clock-regulation of endogenous
CatA mRNA accumulation.
3.4. Expression of the CatA and GUS gene at the pre-mRNA le6el
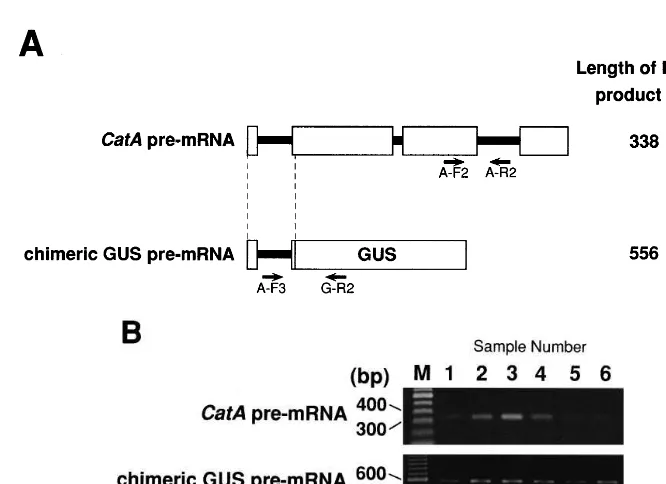
To examine whether the accumulation of pre-mRNAs, which are unprocessed transcripts in nu-clei, for the CatA and the reporter gene show the same patterns as those of mature mRNAs, RT-PCR was performed using DNase I-treated total RNA extracted from the transgenic rice plants. A-F2 and A-R2 primers were used to amplify a region spanning from exon-3 to intron-3 of CatA
pre-mRNA, and A-F3 and G-R2 primers to am-plify a fragment of the chimeric GUS pre-mRNA containing the CatA intron-1 (Fig. 5A). Agarose
Fig. 3. Diurnal oscillations inCatCmRNA abundance. Growth conditions and harvesting time are as described in Section 2 and in the legend to Fig. 2. Northern blot analyses ofCatCmRNA in the leaf blade (A) and leaf sheath (C) were performed using
Fig. 4. Expression of the GUS gene fused with the 1.9-kb fragment ofCatAat the 5’ end. Northern blot analyses of the GUS and the CatA mRNAs in the leaf sheath were per-formed using GUS gene- and CatA-specific probes, respec-tively. Dark and light periods are indicated by black and white bars, respectively, below the figure. The sample number and the time of sample harvest in hours (hr) are indicated above and below the figure, respectively.
mRNA accumulation was very similar to that of the GUS mRNA accumulation (Fig. 4). To verify the absence of contaminating DNA, PCR was performed using DNase I-treated total RNA as a template without reverse transcriptase. Agarose gel electrophoresis showed that no PCR product was amplified, indicating the absence of DNA in the template (data not shown).
4. Discussion
To understand the mode of expression of the three catalase genes at the mRNA level in rice, we performed Northern analysis using total RNAs from various tissues and from plants at various sampling points under LD, LL and DD condi-tions. The CatA mRNA is abundant in the leaf sheath of seedlings and in the leaf blade and flower in mature plants (Fig. 1). In seedlings, leaf sheaths are elongating and do not store surplus assimilates until they are completely elongated [18]. The difference in storing function of leaf sheath between seedlings and mature plants may gel electrophoresis showed that RT-PCR products
of 350 and 550 bp were amplified, each corre-sponding to the fragments of CatA (338 bp) and chimeric GUS (556 bp) pre-mRNAs, respectively (Fig. 5B). The abundance of CatA pre-mRNA showed diurnal oscillations identical to that of
CatA mRNA, and the pattern of the GUS
pre-Fig. 5. Levels of the endogenousCatAand chimeric GUS pre-mRNAs in transgenic plants detected by RT-PCR. (A) Diagram showing the positions of primers (arrows) used for RT-PCR on theCatAand chimeric GUS pre-mRNAs and the expected sizes of the RT-PCR products. Exons and introns are indicated by boxes and thick lines, respectively. Common regions between two pre-mRNAs are shown with dotted lines. (B) Agarose gel electrophoresis of the RT-PCR products showing the abundance of
be related to the CatA expression. In the leaf sheath of seedlings, the CatA mRNA level fluctu-ates with a peak late in the light period (Fig. 2). Since the CatA ortholog in maize, Cat3, is re-ported to be associated with mitochondria [19], the
CatA catalase may also be involved in the decom-position of H2O2 produced in mitochondria at
night. The CatB mRNA is expressed strongly in the root of seedlings, but weakly in the root of mature plants (Fig. 1). This may be a reflection of metabolic changes with maturity. For instance, it is known that the respiration rate is high in young root cells and low in old ones [20]. A high level of
CatC mRNA expression is detected in the leaf blade in seedling and mature plants (Fig. 1), and early in the light period (Fig. 3). The CatC cata-lase may be involved in scavenging the H2O2
gen-erated during photorespiration in the day time. The relation between the abundance of each mRNA species observed here and catalase protein and activity levels remains to be elucidated.
It has been reported that the CatA transcript was highly expressed in the shoot of seedlings and seeds sampled during 0 –+25 DAP [11], and that
CatA was abundant both in the leaf blade and in the leaf sheath at the three-leaf stage [21]. The difference in theCatAmRNA abundance between the leaf blade and the leaf sheath of seedlings was however more prominent in this study than that reported by Chen et al. (1997) [21]. This may indicate either that the accumulation of CatA
transcript varies to some extent among rice strains, or a different sampling time because the abun-dance ofCatAmRNA fluctuates in a day (Fig. 2). Clear daily oscillations of CatC mRNA abun-dance were detected in the leaf blade under DD but not LD and LL conditions (Fig. 3). It seems that these fluctuations are light-independent in contrast to those of genes associated with photo-synthetic organs [22]. Therefore, the expression of
CatC mRNA under LD conditions is supposed to show two patterns: one identical to the clear daily oscillations under DD conditions and the other with a peak late in the dark period and early in the light period. The transcript of tomato biotin-bind-ing protein of unknown function also showed diurnal oscillations only in DD conditions [23]. Our previous study showed that the rice CatA,
CatB and CatC have an exon-intron structure similar to that of maize Cat3, Cat1 and Cat2, respectively, and suggested that three catalase
genes were present in the common ancestor of rice and maize [12]. The tissue-specific expression of the rice CatA, CatB and CatCis similar to that of the maize Cat3, Cat1 and Cat2, respectively. Fur-thermore, expression under a circadian rhythm is also detected in the maizeCat3, and the pattern of diurnal oscillations in the Cat3 mRNA [10] is similar to that in rice CatA mRNA shown in this study. These results suggest that most of the char-acteristics of catalase gene expression were estab-lished in the common ancestor of rice and maize. The expression pattern of the GUS mRNA was not the same as that of CatA mRNA under LD conditions in transgenic plants which have the GUS gene driven by the 5%-flanking region of
CatA (Fig. 4). In addition, the diurnal oscillations of GUS pre-mRNA were not identical to those of mature CatA mRNA, while those of the endoge-nous CatA pmRNA were (Fig. 5B). The re-porter genes driven by the 5%-flanking region of
other genes regulated by a circadian rhythm in higher plants, however, exhibited oscillations of mRNA abundance very similar to those of native gene transcripts in transgenic plants [24 – 27]. For example, the expression of chimeric GUS gene showed diurnal oscillations in transgenic tabacco plants when the promoter region of either of two light-harvesting complex protein genes, wheat Cab-1 [24] or tomato Lhc a4 [27] gene, was fused to the GUS gene. For the circadian regulation of
CatA expression, additional regions such as the
CatA exons, introns and 3%-flanking region, which
have not been fused to the GUS gene, may be crucial. It is worth noting that the 3% untranslated
region of the legume early nodulin gene conferred tissue-specific expression of the GUS reporter gene [28]. From our results, the possibility that the circadian rhythmic expression of CatAis regulated by the degradation of mature mRNA can be ex-cluded. It remains to be seen whether the circadian control of the rice CatAexpression is conferred by transcriptional regulation or by post-transcrip-tional regulation such as modulation of pre-mRNA stability.
Acknowledgements
References
[1] M.L. Abler, J.G. Scandalios, Isolation and characteriza-tion of a genomic sequence encoding the maize Cat3 catalase gene, Plant Mol. Biol. 22 (1993) 1031 – 1038. [2] L. Guan, J.G. Scandalios, Characterization of the
cata-lase antioxidant defense gene Cat1 of maize, and its developmentally regulated expression in transgenic to-bacco, Plant J. 3 (1993) 527 – 536.
[3] L. Guan, A.N. Polidoros, J.G. Scandalios, Isolation, characterization and expression of the maize Cat2 cata-lase gene, Plant Mol. Biol. 30 (1996) 913 – 924.
[4] J.A. Frugoli, H.H. Zhong, M.L. Nuccio, P. McCourt, M.A. McPeek, T.L. Thomas, et al., Catalase is encoded by a multigene family in Arabidopsis thaliana (L.), Heynh. Plant Physiol. 112 (1996) 327 – 336.
[5] H. Willekens, R. Villarroel, M. Van Montagu, D. Inze´, W. Van Camp, Molecular identification of catalases from
Nicotiana plumbaginifolia (L.), FEBS Lett. 352 (1994) 79 – 83.
[6] M. Esaka, N. Yamada, M. Kitabayashi, Y. Setoguchi, R. Tsugeki, M. Kondo, et al., cDNA cloning and differ-ential gene expression of three catalases in pumpkin, Plant Mol. Biol. 33 (1997) 141 – 155.
[7] C.R. McClung, Regulation of catalases in Arabidopsis, Free Radic. Biol. Med. 23 (1997) 489 – 496.
[8] M.G. Redinbaugh, M. Sabre, J.G. Scandalios, The dis-tribution of catalase activity, isozyme protein, and tran-script in the tissues of the developing maize seedling, Plant Physiol. 92 (1990) 375 – 380.
[9] H. Willekens, C. Langebartels, C. Tire´, M. Van Mon-tagu, D. Inze´, W. Van Camp, Differential expression of catalase genes in Nicotiana plumbaginifolia (L.), Proc. Natl. Acad. Sci. USA 91 (1994) 10450 – 10454.
[10] M.G. Redinbaugh, M. Sabre, J.G. Scandalios, Expres-sion of the maize Cat3 catalase gene is under the influ-ence of a circadian rhythm, Proc. Natl. Acad. Sci. USA 87 (1990) 6853 – 6857.
[11] K. Higo, H. Higo, Cloning and characterization of the riceCatAcatalase gene, a homologue of the maizeCat3 gene, Plant Mol. Biol. 30 (1996) 505 – 521.
[12] M. Iwamoto, M. Maekawa, A. Saito, H. Higo, K. Higo, Evolutionary relationship of plant catalase genes inferred from exon-intron structures: isozyme divergence after the separation of monocots and dicots, Theor. Appl. Genet. 97 (1998) 9 – 19.
[13] Y. Tada, M. Sakamoto, T. Fujimura, Efficient gene introduction into rice by electroporation and analysis of transgenic plants: use of electroporation buffer lacking chloride ions, Theor. Appl. Genet. 80 (1990) 475 – 480. [14] A.P. Feinberg, B. Vogelstein, A technique for
radiolabel-ing DNA restriction endonuclease fragments to high specific activity, Anal. Biochem. 132 (1983) 6 – 13. [15] B. Piechulla, ‘Circadian clock’ directs the expression of
plant genes, Plant Mol. Biol. 22 (1993) 533 – 542.
[16] E. Fejes, F. Nagy, Molecular analysis of circadian clock-regulated gene expression in plants: features of the ‘out-put’ pathways, in: P.J. Lumsden, A.J. Millar (Eds.), Biological Rhythms and Photoperiodism in Plants, BIOS Scientific Publishers, Oxford, 1988, pp. 99 – 118. [17] H.H. Zhong, A.S. Resnick, M. Straume, C.R. McClung,
Effects of synergistic signaling by phytochrome A and cryptochrome 1 on circadian clock-regulated catalase expression, Plant Cell 9 (1997) 947 – 955.
[18] T. Mizuochi, Development and aging of leaves, in: T. Matsuo, K. Kumazawa, R. Ishii, K. Ishihara, H. Hirata (Eds.), Science of the Rice Plant, vol. 2, Physiology, Food and Agriculture Policy Research Center, Tokyo, Japan, 1995, pp. 140 – 151.
[19] J.G. Scandalios, W.F. Tong, D.G. Roupakias, Cat3, a third gene locus coding for a tissue-specific catalase in maize: genetics, intracellular location, and some bio-chemical properties, Mol. Gen. Genet. 179 (1980) 33 – 41. [20] R. Ishii, Organ structure and respiration, in: T. Matsuo, K. Kumazawa, R. Ishii, K. Ishihara, H. Hirata (Eds.), Science of the Rice Plant, vol. 2, Physiology, Food and Agriculture Policy Research Center, Tokyo, Japan, 1995, pp. 530 – 531.
[21] Z. Chen, S. Iyer, A. Caplan, D.F. Klessig, B. Fan, Differential accumulation of salicylic acid and salicylic acid-sensitive catalase in different rice tissues, Plant Physiol. 114 (1997) 193 – 201.
[22] C.H. Johnson, M. Knight, A. Trewavas, T. Kondo, A clockwork green: circadian programs in photosynthetic organisms, in: P.J. Lumsden, A.J. Millar (Eds.), Biologi-cal Rhythms and Photoperiodism in Plants, BIOS Scien-tific Publishers, Oxford, 1988, pp. 1 – 34.
[23] G. Giuliano, N.E. Hoffman, K. Ko, P.A. Scolnik, A.R. Cashmore, A light-entrained circadian clock controls transcription of several plant genes, EMBO J. 7 (1988) 3635 – 3642.
[24] E. Fejes, A. Pay, I. Kanevsky, M. Szell, E. Adam, S. Kay, et al., A 268 bp upstream sequence mediates the circadian clock-regulated transcription of the wheatCab -1 gene in transgenic plants, Plant Mol. Biol. -15 (-1990) 921 – 932.
[25] A.J. Millar, S.A. Kay, Circadian control of cab gene transcription and mRNA accumulation in Arabidopsis, Plant Cell 3 (1991) 541 – 550.
[26] Z. Liu, C.C. Taub, C.R. McClung, Identification of an
Arabidopsis thaliana ribulose-1,5-bisphosphate carboxy-lase/oxygenase activase (RCA) minimal promoter regu-lated by light and the circadian clock, Plant Physiol. 112 (1996) 43 – 51.
[27] B. Piechulla, N. Merforth, B. Rudolph, Identification of tomato Lhc promoter regions for circadian expression, Plant Mol. Biol. 38 (1998) 655 – 662.
[28] R. Chen, D.L. Silver, F.J. de Bruijn, Nodule par-enchyma-specific expression of the Seebania rostrata
early nodulin geneSrEnod2 is mediated by its 3% untrans-lated region, Plant Cell 10 (1998) 1585 – 1602.