SBMLChecker, a Semantic approach for SBML model reliability evaluation.
Mathialakan Thavappiragasam, Carol M. Lushbough, Etienne Z. GnimpiebaComputer Science Department, University of South Dakota, 414 E. Clark St. Vermillion, SD 57069, USA, {Mathialakan.Thavappi; Carol.Lushbough; Etienne.Gnimpieba}@usd.edu
ABSTRACT
In Systems Biology model design, reliability evaluation constitutes a requirements challenge. In order to apply the models on a given process or on work for in silico study, a systems biologist needs to be ensured of the models quality. The key problem remains the relation between the model and the biologist question. Several algorithms was designed to validate models but they only check correctness of syntax (e.g. Online SBML validator). These algorithms do not consider semantic annotation of a model defining biological context of the model. In our approach we have measured the model reliability using a combination of meaning (semantic) and syntax. This approach allows researcher to identify a model that really fits his needs and application domain. It also provides unique identification to each model element (compound, reaction, and compartment) in order to facilitate any Systems Biology operation such as merging, splitting, and simulation. It is implemented in Java and connected to the model database BIOMODELS using Restful API, our algorithm implementation called SBMLChecker is
available online at
http://jacksons.usd.edu/SBMLC/. The command line version has been deployed on BioExtract (SBML.org), it remains lacking in many aspects in order to provide the appropriate model in the right context. The reliability of a model depends considerably on the context related to the model design. The development of semantic annotation of biological elements allows systems biologists to connect design context (domain ontology) to a model.
Semantic in biological modeling
There are several organizations (EBI [2], NCBI [3]) maintaining databases ( Biomodels [2], Protein, Gene, etc.) and/or ontologies (gene ontology [4]) in order to manage biological components (e.g., reaction, species, etc.) in a standard way. They try to categorize the already defined components and identify relationships among them. Each database assigns unique id to each element and keeps tracking relevant details (e.g. properties, description) with these ids. Furthermore, we can find several web applications (e.g., KEGG Mapper) that provide services to map the same components from different places [5]. Some of them provide web services, especially RESTful services, that could be used by software tool developers (web services for GO terms and annotation provided by EBML-EBI) [6]. A single component can be annotated by multiple databases and or ontologies. The SBML defines annotation-tag to annotate biological components, it has resources with the details of database and id for each annotation [7]. E.g. the reaction MTHFR, [5,10-methylene-tetrahydrofolate] + [NADPH] → [5-methyl-tetrahydrofolate] in BIOMD0000000018 has the annotations "urn:miriam:ec-code:1.5.1.20", "urn:miriam:kegg.reaction:R01224". This reaction has Enzyme id 1.5.1.20 and KEGG reaction id is R01224.
SBML reliability evaluation in existing tools (Online SBML Validator)
The model reliability checking should ensure their correctness on both syntax and semantic (meaning). The Online SBML validator introduced by SBML.org provides the services to test syntax and internal consistency of an SBML model. This system checks the following aspects of a model [1]:
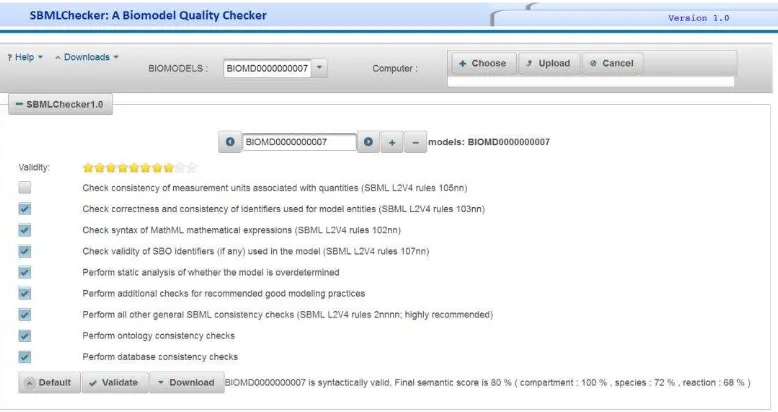
Consistency of measurement units associated with quantities (SBML L2V4 rules 105nn)
Correctness and consistency of identifiers used for model entities (SBML L2V4 rules 103nn)
Validity of SBO identifiers (if any) used in the model (SBML L2V4 rules 107nn)
Perform static analysis of whether the model is over determined
Perform additional checks for recommended good modeling practices
Perform all other general SBML consistency checks (SBML L2V4 rules 2nnnn; highly recommended)
However, this system does not consider the entire annotation information to evaluate the meaning of models. In order to analyze the semantics and syntax of models, we have designed a tool that extends the web services provided by the online SBML validator
2. METHOD
Principle
The reliability level of a model is calculated based on its validity of its syntax and semantics. Correctness of models on syntax is examined with the usage of web services provided by the online SBML validator. Semantic strength is measured by the annotated URL id of each model’s component.
Design and algorithms for SBMLChecker
SBMLChecker does two way analysis, one for semantic strength and another for syntax correctness (Figure 5.), and generates reports R1, R2 respectively.
Figure 1. Global mechanism for model reliability evaluation
Figure 2. Algorithm for semantic analysis
The semantic analyzer takes each kind of component separately and identifies all ontologies and databases that are used to annotate it. For example, if any species is annotated with KEGG id, KEGG will be used to check the annotation of every remaining species. Then the percentage of KEGG annotation will be calculated. Species are considered to be more consistent/reliable if the percentage is high. In this way, percentage of every possible annotated ontologies/databases will be calculated. The maximum percentage will decide the best consistency level of the model element (e.g. species) in the resource (e.g. KEGG, MIRIAM Register)
Reliability score estimation
The consistency for the components ( ) of kind k over ontologies and/or databases,
))
where is an ith ontology or database.
Finally, cumulative consistency is calculated by taking the average consistencies of each kind of component. Consistency of model m,
∑
where the number of components, ∑ . In addition to the consistency report, an error report is generated by combining the online SBML
validator’s error report with our own semantic check error report. Based on the quality checking, it will suggest to provide a valid model for any relevant applications such as model comparison and integration, but it can be skipped if they want.
Implementation of SBMLChecker
We used the IDE NetBeans (7.3) to fulfill everything related to coding, and the JSBML library (jsbml-0.8-with-depenedencies) was used to manipulate SBML files [8]. The JDK 1.7 java library were used for this development [9]. Furthermore, the library to handle excel file: apache poi, and any other relevant libraries were included.
plugins for end user applications, as well as ease migration from a libSBMLbased backend.
Validation
The model BIOMD0000000018 is examined for reliability by SBMLChecker. According to the results, it is syntactically valid and earned a semantic score of 79%. This semantic score comes from: compartment 100%, species 93%, and
reaction 44%. The model’s reliability can be
improved by annotating it.
3. APPLICATION
SBMLChecker in a Workflow Management System (bioextract.org)
Figure 3 SBMLChecker on BioExtract server for the reliability checking of biomodels
A Java program named SBMLChecker.jar is designed for reliability checking. This can process SBML files received through command line parameter argument, and writes a generated report in excel, text, and xml formats. The SBMLChecker has been deployed on the HPC (High Performance
Computing) infrastructure iPlant for availability on BioExtract server (Figure 3), and has been integrated on a web portal (Figure 4).
4. CONCLUSION
SBMLChecker provide a novel approach in SBML model reliability measurement. Using a combination of the meaning (semantic) and the syntax, we generate a reliability score that can be used as indicator to interpret the output result from a given model in specific context. This approach also provides a unique identification to each model element (compound, reaction, and compartment) in order to facilitate any Systems Biology operation
such as merging, splitting, simulation.
Implemented in Java and connected to the model database BIOMODELS using Restful API, SBMLChecker is available online for small models and available on Bioextract.org for workflow design and big models.
Funding: This work was made possible by SD-INBRE Grant #P20RR016479-09 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
REFERENCES
Available: http://www.ncbi.nlm.nih.gov/. [Accessed: 10-Mar-2014]. Bouguerleret, E. Boutet, L. Breuza, A. Bridge, W. M.
Chan, G. Chavali, E. Coudert, E. Dimmer, A. Estreicher, L. Famiglietti, M. Feuermann, A. Gos, N. Gruaz-Gumowski, R. Hieta, C. Hinz, C. Hulo, R. Huntley, J. James, F. Jungo, G. Keller, K. Laiho, D. Legge, P. Lemercier, D. Lieberherr, M. Magrane, M. J. Martin, P. Masson, P. Mutowo-Muellenet, C. Talmud, M. Chibucos, M. G. Giglio, H.-Y. Chang, S. Hunter, C. McAnulla, A. Mitchell, A. Sangrador, R.
Matthews, R. Balakrishnan, G. Binkley, J. M. Cherry, M. C. Costanzo, S. S. Dwight, S. R. Engel, D. G. Ontology annotations and resources.,” Nucleic Acids Res., vol. 41, no. Database issue, pp. D530–5, Jan. 2013.
[5] M. Kanehisa, S. Goto, Y. Sato, M. Furumichi, and M.
Tanabe, “KEGG for integration and interpretation of
large-scale molecular data sets.,” Nucleic Acids Res., vol. 40, no. Database issue, pp. D109–14, Jan. 2012.
Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 1 Core,”
Nat. Preced., Oct. 2010.
[9] O. Corporation, “JavaTM Platform, Standard Edition 7
Development Kit.” [Online]. Available: